
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,123,591 – 21,124,029 |
| Length | 438 |
| Max. P | 0.998324 |

| Location | 21,123,591 – 21,123,711 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
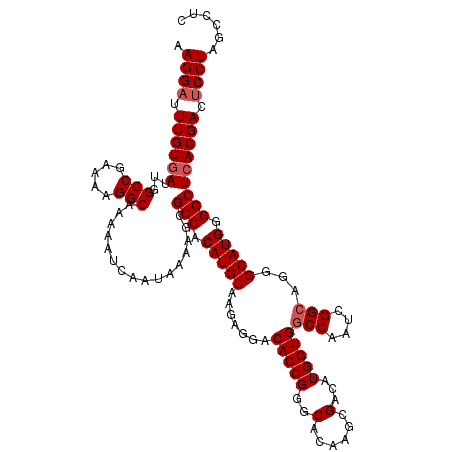
>3L_DroMel_CAF1 21123591 120 - 23771897 AAGGAUUUGUUAUUGGCCGAAAAGGCAAAAAUCAAUAAAAACGCCACAUGUAAGAGGACACCGGACACAAGCGACAUGGUGUCCAAUCGGCAGGGCAUGGGGCUCACGACACCUAGACUC .(((..((((.....(((((...(((................)))..........((((((((..(......)...))))))))..))))).((((.....))))))))..)))...... ( -33.69) >DroSec_CAF1 174298 120 - 1 AAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAAGCGCCACAUGUAAGAGGACACCGGGCACAAGCGACAUGGUGGCCAAUCGGCAGGGCAUGGGGCUCACGACUCCUAGCCUC .((((.((((((...(((.....)))................(((.(((((.......(((((.((....))....)))))(((....)))...))))).))))))))).))))...... ( -39.70) >DroSim_CAF1 85996 120 - 1 AAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAAGCGCCACAUGUAAGAGGACACCGGGCACAAGCGACAUGGUGGCCAAUCGGCAGGGCAUGGGGCUCACGACUCCUAGCCUC .((((.((((((...(((.....)))................(((.(((((.......(((((.((....))....)))))(((....)))...))))).))))))))).))))...... ( -39.70) >DroEre_CAF1 96719 120 - 1 AAGGAUUUGUGAUUGGCCGAAACGGCAAAAAUCAAUAAAAGCGCCACAUGUAAGAGGACACCGGGCACAAGCGACAUGGUGGCCAAUCGGCAGGGCAUGGGGCUCACGACUCCUAGCCUC .((((.((((((...((((...))))................(((.(((((.......(((((.((....))....)))))(((....)))...))))).))))))))).))))...... ( -39.80) >DroYak_CAF1 67686 120 - 1 AAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAAGCGCCACAUGUAAGAGGACACCGGGCACAACCGACAUGGUGGCCAAUCGGCAGGGCAUGGGGCUCACGACUCCUAGCCUC .((((.((((((...(((.....)))................(((.(((((.......(((((..(......)...)))))(((....)))...))))).))))))))).))))...... ( -38.60) >consensus AAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAAGCGCCACAUGUAAGAGGACACCGGGCACAAGCGACAUGGUGGCCAAUCGGCAGGGCAUGGGGCUCACGACUCCUAGCCUC .((((.((((((...(((.....)))................(((.(((((.......(((((..(......)...)))))(((....)))...))))).))))))))).))))...... (-35.10 = -35.70 + 0.60)


| Location | 21,123,671 – 21,123,791 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123671 120 - 23771897 GUGUUGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAAAGGCAAGGGAAGCCGAAGGAUUUGUUAUUGGCCGAAAAGGCAAAAAUCAAUAAAA (((((.((((......)))))))))............(((.....((((((((((((..(......(((.......)))...)..))))))..))))))....))).............. ( -29.50) >DroSec_CAF1 174378 120 - 1 GUGUGGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAAAAGCAAGGGAAGCCGAAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAA ...........((((((((((.(((((((....(((..((..((.((.((......))....))...))...))..))).....))))))).)))(((.....)))...))))))).... ( -28.20) >DroSim_CAF1 86076 120 - 1 GUGUGGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAAAAGCAAGGGAAGCCGAAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAA ...........((((((((((.(((((((....(((..((..((.((.((......))....))...))...))..))).....))))))).)))(((.....)))...))))))).... ( -28.20) >DroEre_CAF1 96799 120 - 1 GUGUGGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAGAAGCAAGGGAAGCCGAAGGAUUUGUGAUUGGCCGAAACGGCAAAAAUCAAUAAAA ...........((((((((((.(((((((....(((..((..((.((.((......))....))...))...))..))).....))))))).)))((((...))))...))))))).... ( -27.20) >DroYak_CAF1 67766 115 - 1 GUGUGGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAAAAGCA-----AGCCGAAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAA ...........((((((((((.(((((((....(((.((((....)))).......(.....).......-----.))).....))))))).)))(((.....)))...))))))).... ( -24.40) >consensus GUGUGGCGAAAAUUGAUUCGAACAUAAAUAAAGGGCAGCCAAGCGUGGCCAGACAAGGACAACAAAAGCAAGGGAAGCCGAAGGAUUUGUGAUUGGCCGAAAAGGCAAAAAUCAAUAAAA ...........((((((((((.(((((((....(((..((..((.((.((......))....))...))...))..))).....))))))).)))(((.....)))...))))))).... (-24.92 = -25.52 + 0.60)


| Location | 21,123,791 – 21,123,911 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -20.12 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123791 120 - 23771897 UGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCAACUGAUAAUAAAGCCACCUCCCUUGCUCACUUCUUCUUUUCAUUCGCUCCAUUCACAGGAGGACAAUAAGAAAACUGCAA ....(((((.(((....(((...((((........))))...)))...))).)))))......(((.......(((((.....((.((((.......))))))....)))))....))). ( -21.50) >DroSec_CAF1 174498 120 - 1 CGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCAACUGAUAAUAAAGCCACCUCCCUUGCUCACUUCUUCUUUUCUUUCGCUCCAUUCACAGGAGGACAAUAAGAAAACUGCAA .(..(((((.(((....(((...((((........))))...)))...))).)))))..)...(((....(((((.((.((((((............)))))).)).)))))....))). ( -22.60) >DroSim_CAF1 86196 120 - 1 UGUCGUGGCAUUAACACUCGCACGGGACAUACUAUUUCCAACUGAUAAUAAAGCCACCUCCCUUGCUCACUUCUUCUUUUCUUUCGCUCCAUUCACAGGAGGACAAUAAGAAAACUGCAA ....(((((.(((....(((...((((........))))...)))...))).)))))......(((....(((((.((.((((((............)))))).)).)))))....))). ( -21.80) >DroEre_CAF1 96919 116 - 1 UGUCGUGGCAUUAACACUCACACGGGACACACUAUUUCCAACUGAUAAUAAAGCCACCUC-CUUGCUCUC---UUCCUUUCUUUCGCUCCAUUCACAGGACGACAAUAAGAAAACUGCAA ....(((((.(((....(((...((((........))))...)))...))).)))))...-..(((....---....(((((((((.(((.......))))))....))))))...))). ( -19.60) >DroYak_CAF1 67881 117 - 1 UGUCGUGGCAUUAGCACUCACACGGGACAUAGUAUUUCCAUCUGAUAAUAAAGCCACAUCCCUUGCUCAC---UUCUUUUCUUUCGCUCCAUUCACAGGAGGACAAUAAGAAAACUGCAA ....(((((.(((....(((...((((........))))...)))...))).)))))......(((....---(((((.....((.((((.......))))))....)))))....))). ( -25.30) >consensus UGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCAACUGAUAAUAAAGCCACCUCCCUUGCUCACUUCUUCUUUUCUUUCGCUCCAUUCACAGGAGGACAAUAAGAAAACUGCAA ....(((((.(((....(((...((((........))))...)))...))).)))))......(((.......(((((.....((.((((.......))))))....)))))....))). (-20.12 = -20.36 + 0.24)


| Location | 21,123,871 – 21,123,991 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123871 120 - 23771897 AAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAAUGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCA ...((((((((((..(.(((((...)))))..)..))))......(((((.((((((.(((.......))).......)))))))))))..............))))))........... ( -31.00) >DroSec_CAF1 174578 120 - 1 AAUUGUCUCAUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAACGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCA ...((((((.......(((....)))((..((((((.(((.....(((...)))....(((.......)))..))).)))))..)..))..............))))))........... ( -27.20) >DroSim_CAF1 86276 120 - 1 AAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAAUGUCGUGGCAUUAACACUCGCACGGGACAUACUAUUUCCA ...((((((((((..(.(((((...)))))..)..))))......(((((.((((((.(((.......))).......)))))))))))..............))))))........... ( -31.00) >DroEre_CAF1 96995 120 - 1 AAUUGUCUCGUUAUUCCUUGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUGGGGCCGUCAAAUGUCGUGGCAUUAACACUCACACGGGACACACUAUUUCCA ...((((((((.......((((((((((.................)))...(((....(((......))).))).........)))))))...........))))))))........... ( -30.90) >DroYak_CAF1 67958 120 - 1 AAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCGUUGGGCCGUCAAAUGUCGUGGCAUUAGCACUCACACGGGACAUAGUAUUUCCA ...((((((((....((((....)))).(((((......(....)(((((.((((((.(((.......))).......)))))))))))..))))).....))))))))........... ( -31.70) >consensus AAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAAUGUCGUGGCAUUAACACUCACACGGGACAUACUAUUUCCA ...((((((((((..(.(((((...)))))..)..))))......(((((.((((((.......((....))......)))))))))))..............))))))........... (-27.74 = -28.18 + 0.44)



| Location | 21,123,911 – 21,124,029 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.90 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123911 118 - 23771897 GGCCCGAUGUUAAUUGAAUUGUCUGCGGCCC--CGAGCGCAAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAA (((((((.(..(((.(...((((...(((..--.(((.((....)))))((((..(.(((((...)))))..)..))))......)))...))))...).)))..).)))))))...... ( -35.80) >DroSec_CAF1 174618 118 - 1 GGCCCGAUGUUAAUUGAAUUGUCUGCGGCCC--CGAGCGCAAUUGUCUCAUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAA (((((((.(..(((.(((((((..(((((..--.(((.((....)))))......((((....))))..)))))...))))).))(((...)))......)))..).)))))))...... ( -34.30) >DroSim_CAF1 86316 118 - 1 GGCCCGAUGUUAAUUGAAUUGUCUGCGGCCC--CGAGCGCAAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAA (((((((.(..(((.(...((((...(((..--.(((.((....)))))((((..(.(((((...)))))..)..))))......)))...))))...).)))..).)))))))...... ( -35.80) >DroEre_CAF1 97035 119 - 1 GGC-CGAUGUUAAUUGAAUUGUCUGCGGCCCUUUGAGCGCAAUUGUCUCGUUAUUCCUUGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUGGGGCCGUCAAA (((-((((((((((((((((((..(((((((...(((.((....)))))((........))...)))))))......))))).)))..))))))))).(((......)))))))...... ( -36.40) >DroYak_CAF1 67998 120 - 1 GGCCCGAUGUUAAUUGAAUUGUCUGCGGUCCUCCGCGCGCAAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCGUUGGGCCGUCAAA (((((((((..(((.(((((((.(((((....))))).))))))...((((((..(.(((((...)))))..)..))))))....(((...)))....).)))..)))))))))...... ( -46.20) >consensus GGCCCGAUGUUAAUUGAAUUGUCUGCGGCCC__CGAGCGCAAUUGUCUCGUUAUUCCCAGCCAUGGGCUGCUGUUUAACGAUCUCGCCAUUGGCAUUACCAUUUCCUUUGGGCCGUCAAA (((((((.(..(((.(...((((...(((.....(((.((....)))))((((..(.(((((...)))))..)..))))......)))...))))...).)))..).)))))))...... (-29.94 = -30.62 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:48 2006