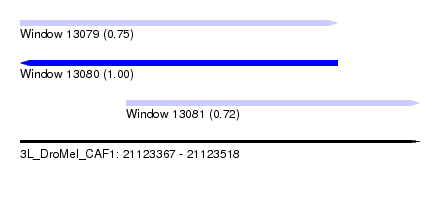
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,123,367 – 21,123,518 |
| Length | 151 |
| Max. P | 0.999090 |

| Location | 21,123,367 – 21,123,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -36.30 |
| Energy contribution | -36.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123367 120 + 23771897 UAGUUAGGCAUGUAAGAAGCACUAGAAGAAGACCCAUAGCCGUAGCCAUAGGUAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGUGACAUUGUCGGCGUAAGAUGU ..(((.(((((((.....((((((..............((((((.(....).))))))..(((.((((((((........)))))))).))).)))))).))).)))))))......... ( -39.20) >DroSec_CAF1 174080 120 + 1 UAGUUAGGCAUGUAAGAAGCACUAGAAGAAGACCCAUAGCCGUAGCCAUAGGGAUGGUAACCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGA ......(((((((.....((((((........(((((.((....)).)).))).......(((.((((((((........)))))))).))).)))))).))).))))..(....).... ( -39.90) >DroSim_CAF1 85778 120 + 1 UAGUUAGGCAUGUAAGAAGCACUAGAAGAAGACCCAUAGCCGUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGA ......(((((((.....((((((........(((((.((....)).)).))).......(((.((((((((........)))))))).))).)))))).))).))))..(....).... ( -39.60) >DroEre_CAF1 96486 120 + 1 UAGUUGGGCAUGUAAGAAGCACCUGCAGAAGACCCAUAUCCAUAGCCAUAGGGAUGGUAGCCGGUGUAGCCCGAUCUGUAGGGCUGCGGCGGAUAGUGCGACAUCGUCGGAUGCGGAUGA ....(((((.((((.(......)))))...).))))(((((...(((((....)))))..(((.((((((((........)))))))).)))...((.((((...)))).))..))))). ( -42.00) >DroYak_CAF1 67455 120 + 1 UAGUUGGGCAUGUAAGAAGCACUAGAAGAAGACCCAUAGCCAUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGACCUGUAGGGCUGCGGCGGAUAGUGCGACAUCGUCGGCGUCUGAUGA ..((..(((.........((((((..............(((((..((...)).)))))..(((.((((.(((........))).)))).))).))))))(((...)))...)))..)).. ( -37.20) >consensus UAGUUAGGCAUGUAAGAAGCACUAGAAGAAGACCCAUAGCCGUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGA ......(((((((.....((((((....................(((((....)))))..(((.((((((((........)))))))).))).)))))).)))).)))............ (-36.30 = -36.50 + 0.20)

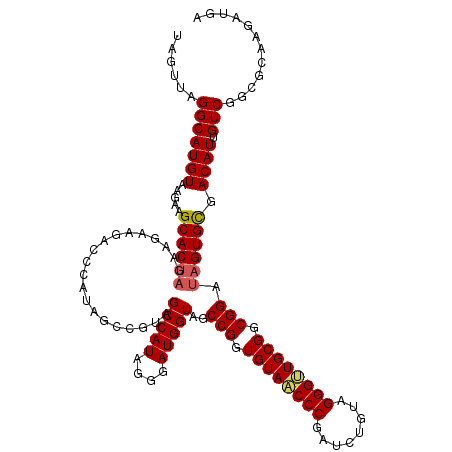
| Location | 21,123,367 – 21,123,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123367 120 - 23771897 ACAUCUUACGCCGACAAUGUCACACUAUCCGCCGCAACCCUACAGAUCGGGUUACACCGGCUACCAUACCUAUGGCUACGGCUAUGGGUCUUCUUCUAGUGCUUCUUACAUGCCUAACUA .....(((.((.(((...))).(((((...((((.(((((........)))))....))))......(((((((((....))))))))).......)))))..........)).)))... ( -32.70) >DroSec_CAF1 174080 120 - 1 UCAUCUUGCGCCGACAAUGUCGCACUAUCCGCCGCAACCCUACAGAUCGGGUUACACCGGUUACCAUCCCUAUGGCUACGGCUAUGGGUCUUCUUCUAGUGCUUCUUACAUGCCUAACUA .......(((..(((...)))((((((...((((.(((((........)))))....)))).......((((((((....))))))))........))))))........)))....... ( -33.60) >DroSim_CAF1 85778 120 - 1 UCAUCUUGCGCCGACAAUGUCGCACUAUCCGCCGCAACCCUACAGAUCGGGUUACACCGGCUACCAUCCCUAUGGCUACGGCUAUGGGUCUUCUUCUAGUGCUUCUUACAUGCCUAACUA .......(((..(((...)))((((((...((((.(((((........)))))....)))).......((((((((....))))))))........))))))........)))....... ( -36.10) >DroEre_CAF1 96486 120 - 1 UCAUCCGCAUCCGACGAUGUCGCACUAUCCGCCGCAGCCCUACAGAUCGGGCUACACCGGCUACCAUCCCUAUGGCUAUGGAUAUGGGUCUUCUGCAGGUGCUUCUUACAUGCCCAACUA ......(((((.(..((.(.(.((.(((((((((.(((((........)))))....))))..((((....))))....))))))).).).))..).))))).................. ( -33.60) >DroYak_CAF1 67455 120 - 1 UCAUCAGACGCCGACGAUGUCGCACUAUCCGCCGCAGCCCUACAGGUCGGGUUACACCGGCUACCAUCCCUAUGGCUAUGGCUAUGGGUCUUCUUCUAGUGCUUCUUACAUGCCCAACUA ......((((.......))))((((((...((((.(((((........)))))....)))).......((((((((....))))))))........)))))).................. ( -33.50) >consensus UCAUCUUACGCCGACAAUGUCGCACUAUCCGCCGCAACCCUACAGAUCGGGUUACACCGGCUACCAUCCCUAUGGCUACGGCUAUGGGUCUUCUUCUAGUGCUUCUUACAUGCCUAACUA ............(((...)))((((((...((((.(((((........)))))....)))).......((((((((....))))))))........)))))).................. (-31.36 = -31.24 + -0.12)

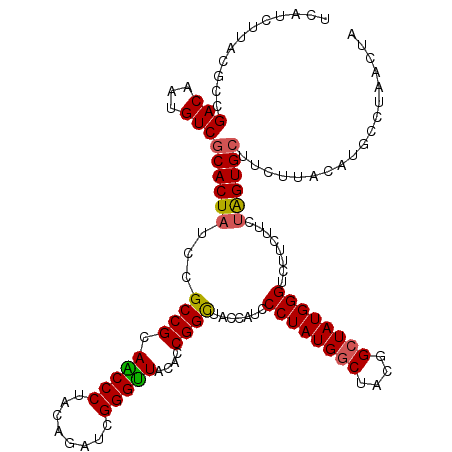
| Location | 21,123,407 – 21,123,518 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -40.34 |
| Consensus MFE | -31.78 |
| Energy contribution | -31.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123407 111 + 23771897 CGUAGCCAUAGGUAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGUGACAUUGUCGGCGUAAGAUGUU---GCGGAUGAUGAU------GACUAUGAUGCUGAUCGU ..(((((((((.((((.((.(((.((((((((........)))))))).))).)).)))).(((..((.(((........)---)).))..)))..------..)))))..))))..... ( -40.50) >DroSec_CAF1 174120 105 + 1 CGUAGCCAUAGGGAUGGUAACCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGAU---GCGGAUGGUGAU------------GAUGCUGAUCGU .((.(((((....))))).))((.((((((((........)))))))).))(((...(((.(((..(((.((((......)---)))..)))..))------------).)))..))).. ( -39.30) >DroSim_CAF1 85818 105 + 1 CGUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGAU---GCGGAUGGUGAU------------GAUGCUGAUCGU .((.(((((....))))).))((.((((((((........)))))))).))(((...(((.(((..(((.((((......)---)))..)))..))------------).)))..))).. ( -39.60) >DroEre_CAF1 96526 120 + 1 CAUAGCCAUAGGGAUGGUAGCCGGUGUAGCCCGAUCUGUAGGGCUGCGGCGGAUAGUGCGACAUCGUCGGAUGCGGAUGAUAUGGCGGAUGGUGCUGACUAUGCUGAUGAUGCUGAUCGU ....(((((....)))))..(((.((((((((........)))))))).))).....(((((((((((((.(((.(......).))).(((((....))))).)))))))))....)))) ( -42.90) >DroYak_CAF1 67495 111 + 1 CAUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGACCUGUAGGGCUGCGGCGGAUAGUGCGACAUCGUCGGCGUCUGAUGAU---GCGGGUGGUGAU------GAUGAUGAUGCUGAUCGU ....(((((....)))))..(((.((((.(((........))).)))).))).(((((((.((((((((.(..(((.....---.)))..).))))------)))).)).)))))..... ( -39.40) >consensus CGUAGCCAUAGGGAUGGUAGCCGGUGUAACCCGAUCUGUAGGGUUGCGGCGGAUAGUGCGACAUUGUCGGCGCAAGAUGAU___GCGGAUGGUGAU______G___AUGAUGCUGAUCGU ....(((((....)))))..(((.((((((((........)))))))).))).........(((((((.((.............)).)))))))..............(((....))).. (-31.78 = -31.38 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:43 2006