
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,122,933 – 21,123,133 |
| Length | 200 |
| Max. P | 0.997330 |

| Location | 21,122,933 – 21,123,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -51.32 |
| Consensus MFE | -45.52 |
| Energy contribution | -44.92 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21122933 120 + 23771897 CGAGCUUUGGGGUUCCAGGCAGCGGGUGAUGUUGCAGCUGCUGUUGGCUCAGAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAAAUAGCUCGGUG ((((((....((..((.((((.(((((.(.((.(((((..(((......)))..))))).))).)))).).))))(((((.((((......)))).)))))))..))....))))))... ( -52.40) >DroSec_CAF1 173646 120 + 1 CGAGCUUUGGGGUUCCAGGCAGCGGGUGAUGUUGCAGCUGCUGUUGGUUCAGAUGCUGCUGUUGACUCAAGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUG .(((((....)))))..(((((((..(((.((((((((..(((......)))..)))))....))))))..)))))))(((((((.(((.((((((...)))))).)))...))))))). ( -47.90) >DroSim_CAF1 85338 120 + 1 CGAGCUUUGGAGUUCCAGGCAGCGGGUGAUGUUGCAGCUGCUGCUGGCUCAGAUGCUGCUGCUGACUAAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAUCGGUUCCAGAUAGCUGGGUG .(((((.(((....)))((((((((.((......)).))))))))))))).(((((.((.((((.....(((((((.....))))))).)))).)))))))...(((((....))))).. ( -51.80) >DroEre_CAF1 96064 120 + 1 CGAGCUUUGGGGUUCCAGGCAGCGGGUGAUGUUGCAGCUGCUGUUGGCUCAAAUGCUGCUGCUGACUGUGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUG .(((((.(((....)))((((((((.((......)).)))))))))))))....(((((((((..(((..((((((.....))))))..)))..).))))))))(((((....))))).. ( -51.50) >DroYak_CAF1 67021 120 + 1 CGAGCUUUGGGGUUCCAGGCAGCGGCUGAUGUUGCAGUUGCUGUUGGCUCAAAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGGUAGCUGGGUG .(((((.(((....)))(((((((((((......))))))))))))))))..((.(.((((((......((..(((((((.((((......)))).)))))))..)).)))))).).)). ( -53.00) >consensus CGAGCUUUGGGGUUCCAGGCAGCGGGUGAUGUUGCAGCUGCUGUUGGCUCAGAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUG (.((((((((((..((((.((((((.((......)).))))))))))..)....((((((((..((....))((.(((((........))))).)))))))))))))))..)))).)... (-45.52 = -44.92 + -0.60)



| Location | 21,122,933 – 21,123,053 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.76 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21122933 120 - 23771897 CACCGAGCUAUUUGGAACCGCUGCGCAGCUACAGCGCCGAGCAGCAACACCUAAGUCAGCAGCAGCAUCUGAGCCAACAGCAGCUGCAACAUCACCCGCUGCCUGGAACCCCAAAGCUCG ...((((((..((((...(((((........)))))(((.(((((.........(....).(((((..(((......)))..)))))..........))))).)))....)))))))))) ( -41.00) >DroSec_CAF1 173646 120 - 1 CACCCAGCUAUCUGGAACCGCUGCGCAGCUACAGCGCCGAGCAGCAACACUUGAGUCAACAGCAGCAUCUGAACCAACAGCAGCUGCAACAUCACCCGCUGCCUGGAACCCCAAAGCUCG .....((((...(((...(((((........)))))(((.(((((......((......))(((((..(((......)))..)))))..........))))).)))....))).)))).. ( -34.40) >DroSim_CAF1 85338 120 - 1 CACCCAGCUAUCUGGAACCGAUGCGCAGCUACAGCGCCGAGCAGCAACACCUUAGUCAGCAGCAGCAUCUGAGCCAGCAGCAGCUGCAACAUCACCCGCUGCCUGGAACUCCAAAGCUCG ...((((....)))).......((((.......))))(((((.((.((......))..)).(((((...(((..((((....)))).....)))...))))).(((....)))..))))) ( -33.90) >DroEre_CAF1 96064 120 - 1 CACCCAGCUAUCUGGAACCGCUGCGCAGCUACAGCGCCGAGCAGCAACACCACAGUCAGCAGCAGCAUUUGAGCCAACAGCAGCUGCAACAUCACCCGCUGCCUGGAACCCCAAAGCUCG .....((((...(((...(((((........)))))(((.(((((.((......))..(((((.((..(((...)))..)).)))))..........))))).)))....))).)))).. ( -33.50) >DroYak_CAF1 67021 120 - 1 CACCCAGCUACCUGGAACCGCUGCGCAGCUACAGCGCCGAGCAGCAACACCUAAGUCAGCAGCAGCAUUUGAGCCAACAGCAACUGCAACAUCAGCCGCUGCCUGGAACCCCAAAGCUCG ...((((....))))....(((((((.((....))((......)).............)).)))))....((((...((((..(((......)))..))))..(((....)))..)))). ( -32.70) >consensus CACCCAGCUAUCUGGAACCGCUGCGCAGCUACAGCGCCGAGCAGCAACACCUAAGUCAGCAGCAGCAUCUGAGCCAACAGCAGCUGCAACAUCACCCGCUGCCUGGAACCCCAAAGCUCG .....((((...(((...(((((........)))))(((.(((((.........(....).(((((..(((......)))..)))))..........))))).)))....))).)))).. (-31.60 = -31.76 + 0.16)



| Location | 21,122,973 – 21,123,093 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -53.94 |
| Consensus MFE | -48.36 |
| Energy contribution | -48.12 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21122973 120 + 23771897 CUGUUGGCUCAGAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAAAUAGCUCGGUGGCAACACGGUGCGAUUUGGAGCGGGCGUGGAGGAUCUGCC .....(((..((((.((.(((((...........((((((.((((......)))).))))))(((((((((.((.(.(((....))).).)).))))))))).)))).).)).))))))) ( -54.00) >DroSec_CAF1 173686 120 + 1 CUGUUGGUUCAGAUGCUGCUGUUGACUCAAGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGCGCUGGCGUCGAGGAUCUGCC ..((.(((((.((((((.............(((((((..((((((.(((.((((((...)))))).)))...))))))..)))))))(((((......)))))))))))..))))).)). ( -56.10) >DroSim_CAF1 85378 120 + 1 CUGCUGGCUCAGAUGCUGCUGCUGACUAAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAUCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGCGCUGGCGUCGAGGAUCUGCC ..((.((((..((((((.............(((((((..((((((.(((.(((((.....))))).)))...))))))..)))))))(((((......)))))))))))..)).)).)). ( -51.70) >DroEre_CAF1 96104 120 + 1 CUGUUGGCUCAAAUGCUGCUGCUGACUGUGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAGCACAGUGCGAUUUGGAGCUGGCGUGGAGGACCUACC ..((..((((...((((((..((.(((((((..(((((((.((((......)))).)))))))..)))..))).).))..))))))(((......)))))))..))((((.....)))). ( -54.70) >DroYak_CAF1 67061 120 + 1 CUGUUGGCUCAAAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGGUAGCUGGGUGGCAACACGGUGCGAUUUGGAGCUGGCGUGGAGGAUCUACC ..((..((((((((((.(((.(((...)))(((((((..((((((.(((.((((((...)))))).)))...))))))..)))))))))))).)))).))))..))((((.....)))). ( -53.20) >consensus CUGUUGGCUCAGAUGCUGCUGCUGACUUAGGUGUUGCUGCUCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGAGCUGGCGUGGAGGAUCUGCC ..((..((((((((((.(((.((.....))(((((((..((((((.(((.((((((...)))))).)))...))))))..)))))))))))).)))))).))..))((((.....)))). (-48.36 = -48.12 + -0.24)

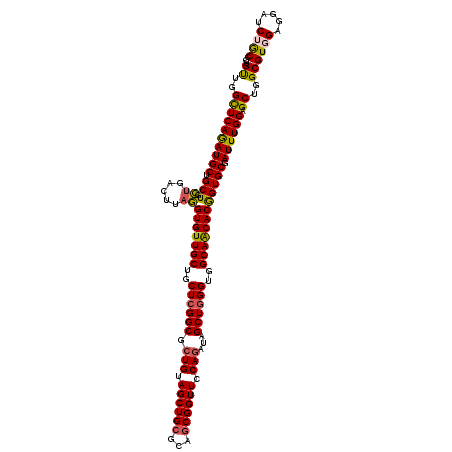
| Location | 21,123,013 – 21,123,133 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -56.58 |
| Consensus MFE | -49.68 |
| Energy contribution | -50.80 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21123013 120 + 23771897 UCGGCGCUGUAGCUGCGCAGCGGUUCCAAAUAGCUCGGUGGCAACACGGUGCGAUUUGGAGCGGGCGUGGAGGAUCUGCCUGGUGGCGCAUAGUGGGGCGUUGGGCCCGCAAAGGCACCG .(((.(((...(((((((.((.(((((((((.((.(.(((....))).).)).)))))))))(((((.(......)))))).)).))))).....((((.....))))))...))).))) ( -56.00) >DroSec_CAF1 173726 120 + 1 UCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGCGCUGGCGUCGAGGAUCUGCCAGGUGGCGCAUAGUGCGGCGUUGGGCCCGCAAAGCCACCG ((((((((((.((..(.((((...........)))).)..)).)))((((((......)))))))))))))((.....)).((((((......(((((.(.....))))))..)))))). ( -57.90) >DroSim_CAF1 85418 120 + 1 UCGGCGCUGUAGCUGCGCAUCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGCGCUGGCGUCGAGGAUCUGCCAGGUGGCGCAUAGUGCGGCGUUGGGCCCGCAAAGCCACCG ((((((((...(((.((((((....((((....))))(((....)))))))))....)))...))))))))((.....)).((((((......(((((.(.....))))))..)))))). ( -57.70) >DroEre_CAF1 96144 120 + 1 UCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAGCACAGUGCGAUUUGGAGCUGGCGUGGAGGACCUACCAGGUGGCGCAUAGUGGGGCGUUGGGCCCGCAAAGCCGCCG ((.(((((...(((....)))((((((((((.((...(((....)))...)).))))))))))))))).))((.....)).((((((.....(((((.(....).)))))...)))))). ( -54.50) >DroYak_CAF1 67101 120 + 1 UCGGCGCUGUAGCUGCGCAGCGGUUCCAGGUAGCUGGGUGGCAACACGGUGCGAUUUGGAGCUGGCGUGGAGGAUCUACCAGGUGGCGCGUAGUGGGGCGUUGGGCCCGCAAAGCCACCA ((.(((((...(((....)))((((((((((.((...(((....)))...)).))))))))))))))).))((.....)).((((((.....(((((.(....).)))))...)))))). ( -56.80) >consensus UCGGCGCUGUAGCUGCGCAGCGGUUCCAGAUAGCUGGGUGGCAACACGGUGCGAUUUGGAGCUGGCGUGGAGGAUCUGCCAGGUGGCGCAUAGUGGGGCGUUGGGCCCGCAAAGCCACCG ..((((...(..((((((..(((((((((((.((...(((....)))...)).)))))))))))))))))..)...)))).((((((.....(((.(((.....))))))...)))))). (-49.68 = -50.80 + 1.12)

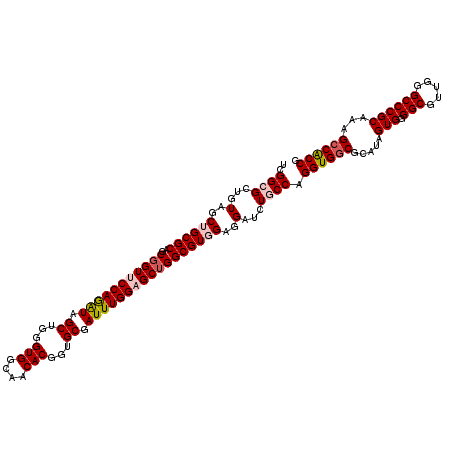

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:40 2006