
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,119,646 – 21,120,001 |
| Length | 355 |
| Max. P | 0.997330 |

| Location | 21,119,646 – 21,119,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -31.76 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997330 |
| Prediction | RNA |
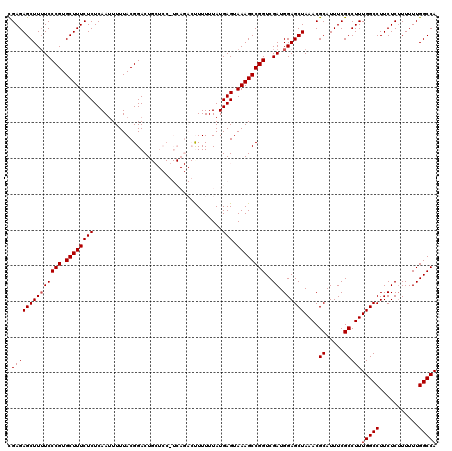
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119646 120 + 23771897 CCCAAAAAUCAAUAGGCGCCCCAUUUUGUGUACUGCUGGAGCACCCUGUAAAAAGGCUUCCGGCUGUGGUUCUCUCCAUUUGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACU .((((((((....((((((...........(((.(((((((...(((......))).))))))).)))((((((.......)))))).......)))))).......))))))..))... ( -29.60) >DroSec_CAF1 170307 120 + 1 CCCAAAAAUCAAUAGGCGCCCCAUUUUGUGUACGGCUGGAGCACCCUGUAAAAAGGCUUCCGGCCGUGGUUCUCUCCAUUCGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACU .((((((((....((((((...........(((((((((((...(((......))).)))))))))))((((((.......)))))).......)))))).......))))))..))... ( -35.10) >DroSim_CAF1 82018 118 + 1 C-CAAAAAUCAAUAGGCGCCCCAUUUUGUGUACGGCUGGAGCACCCUGUAAAAAGGCUUCCGGCCGUGGUUCUCUCCAUUCGAGAGCUU-UCCCGUGCUUUCUCUCAAUUUUUACGGACU (-(((((((....((((((...........(((((((((((...(((......))).)))))))))))((((((.......))))))..-....)))))).......))))))..))... ( -34.80) >DroEre_CAF1 92796 120 + 1 CCCAAAAAUCAAUAGGCGCCCCAUUUUAUGUACGGCUGGAGCACCCUGUAAAAAGGCUUCCGGCCGUGGUUCUCUCCAUUCGAGAGCUUUUCCCGUGCUUUCUCUCAAAUUUUACGGACU .((.(((((....((((((...........(((((((((((...(((......))).)))))))))))((((((.......)))))).......))))))........)))))..))... ( -33.90) >DroYak_CAF1 63701 120 + 1 UCCAAAAAUCAAUAGGCGCCCCAUUUUAUGUACGGCUGGAACACCCUGUAAAAAGGCUUCCGGCCGCGGUUCUCUCCAUUCGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACU (((((((((....((((((.............(((((((((...(((......))).))))))))).(((((((.......)))))))......)))))).......))))))..))).. ( -34.40) >consensus CCCAAAAAUCAAUAGGCGCCCCAUUUUGUGUACGGCUGGAGCACCCUGUAAAAAGGCUUCCGGCCGUGGUUCUCUCCAUUCGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACU .............((((((...........(((((((((((...(((......))).)))))))))))((((((.......)))))).......)))))).................... (-31.76 = -32.00 + 0.24)



| Location | 21,119,646 – 21,119,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119646 120 - 23771897 AGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCAAAUGGAGAGAACCACAGCCGGAAGCCUUUUUACAGGGUGCUCCAGCAGUACACAAAAUGGGGCGCCUAUUGAUUUUUGGG ...((.(((((((..((((((.((.(((......(((((.....)))))........)))...)).)))))...((((((((((.............)))))))))).)..))))))))) ( -35.86) >DroSec_CAF1 170307 120 - 1 AGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCGAAUGGAGAGAACCACGGCCGGAAGCCUUUUUACAGGGUGCUCCAGCCGUACACAAAAUGGGGCGCCUAUUGAUUUUUGGG ...((.(((((((..(.((...((..((......(((((.....)))))..))(((((.(((.(((((.....)))))..))).)))))............))..)).)..))))))))) ( -36.20) >DroSim_CAF1 82018 118 - 1 AGUCCGUAAAAAUUGAGAGAAAGCACGGGA-AAGCUCUCGAAUGGAGAGAACCACGGCCGGAAGCCUUUUUACAGGGUGCUCCAGCCGUACACAAAAUGGGGCGCCUAUUGAUUUUUG-G ......(((((((..(.((...((..((..-...(((((.....)))))..))(((((.(((.(((((.....)))))..))).)))))............))..)).)..)))))))-. ( -35.60) >DroEre_CAF1 92796 120 - 1 AGUCCGUAAAAUUUGAGAGAAAGCACGGGAAAAGCUCUCGAAUGGAGAGAACCACGGCCGGAAGCCUUUUUACAGGGUGCUCCAGCCGUACAUAAAAUGGGGCGCCUAUUGAUUUUUGGG .((((.....(((((((((....(....).....)))))))))((......))(((((.(((.(((((.....)))))..))).)))))..........)))).((((........)))) ( -36.00) >DroYak_CAF1 63701 120 - 1 AGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCGAAUGGAGAGAACCGCGGCCGGAAGCCUUUUUACAGGGUGUUCCAGCCGUACAUAAAAUGGGGCGCCUAUUGAUUUUUGGA ..((((..(((((..(.((...((..((......(((((.....)))))..))(((((.(((((((((.....))))).)))).)))))............))..)).)..))))))))) ( -35.70) >consensus AGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCGAAUGGAGAGAACCACGGCCGGAAGCCUUUUUACAGGGUGCUCCAGCCGUACACAAAAUGGGGCGCCUAUUGAUUUUUGGG ......(((((((..((((((.((.(((......(((((.....)))))........)))...)).)))))...((((((((((.............)))))))))).)..))))))).. (-33.72 = -33.76 + 0.04)

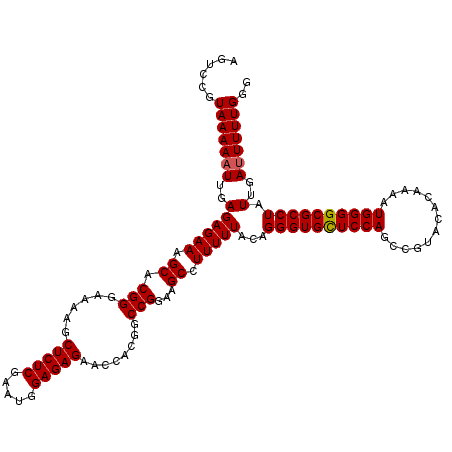

| Location | 21,119,726 – 21,119,846 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119726 120 + 23771897 UGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACUGCUCCGUCAGACUUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCA .((((((.........))))))............((....((((((((.((((.(((((....)))))..))))))))))))....))...........(((((...........))))) ( -33.30) >DroSec_CAF1 170387 108 + 1 CGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACU------------UUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCA .((((((.........)))))).............((.((------------((..........))))))((((((.(((((((((.((.....)).))))))....)))...)))))). ( -26.20) >DroSim_CAF1 82097 107 + 1 CGAGAGCUU-UCCCGUGCUUUCUCUCAAUUUUUACGGACU------------UUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCA .((((((..-......)))))).............((.((------------((..........))))))((((((.(((((((((.((.....)).))))))....)))...)))))). ( -27.00) >DroEre_CAF1 92876 120 + 1 CGAGAGCUUUUCCCGUGCUUUCUCUCAAAUUUUACGGACUGCUCCCUCAGACCUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCA (((((((.....(((((...............)))))...(((((.((.((((.(((((....)))))..)))))).))))).....)).)))))....(((((...........))))) ( -32.36) >DroYak_CAF1 63781 120 + 1 CGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACUGCUCCGUCAGACCUUUUUAUGAGGAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCAUUUUUGGCCA (((((((.....(((((...............)))))...((((((((.(((((((((.....)))))..)))))))))))).....)).)))))....(((((...........))))) ( -35.96) >consensus CGAGAGCUUUUCCCGUGCUUUCUCUCAAUUUUUACGGACUGCUCC_UCAGACUUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCA .((((((((((((((.((((((((....................................))).))))))))..)).))))))....((.....)))))(((((...........))))) (-23.38 = -23.58 + 0.20)

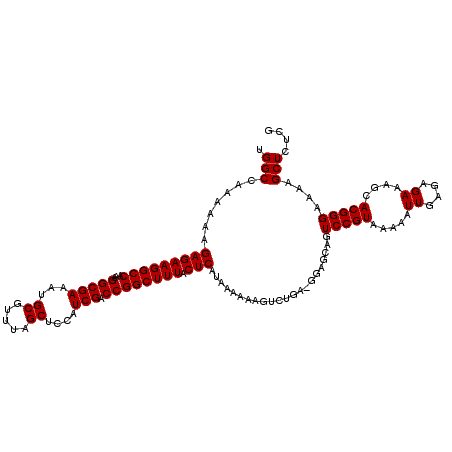
| Location | 21,119,726 – 21,119,846 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119726 120 - 23771897 UGGCCAAAAAAGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUACUCAUAAAAAAGUCUGACGGAGCAGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCA (((((...........))))).........((((..(.(((((.(((...((((((.........))))))))).))))).)..)))).....((((((....(....).....)))))) ( -31.00) >DroSec_CAF1 170387 108 - 1 UGGCCAAAAAAGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUACUCAUAAAAAA------------AGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCG (((((...........)))))..(((((...((.....))....))).)).(((((.((((.......------------.............))))..))))).(((((.....))))) ( -25.05) >DroSim_CAF1 82097 107 - 1 UGGCCAAAAAAGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUACUCAUAAAAAA------------AGUCCGUAAAAAUUGAGAGAAAGCACGGGA-AAGCUCUCG (((((...........)))))..(((((...((.....))....))).)).(((((.((((.......------------.............))))..))))).(((((-....))))) ( -25.05) >DroEre_CAF1 92876 120 - 1 UGGCCAAAAAAGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUACUCAUAAAAAGGUCUGAGGGAGCAGUCCGUAAAAUUUGAGAGAAAGCACGGGAAAAGCUCUCG .((((......(((((((((...(((((...((.....))....))).)))))))).))).......))))...((((((..(((((....(((....)))...)))))....)))))). ( -36.42) >DroYak_CAF1 63781 120 - 1 UGGCCAAAAAUGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUCCUCAUAAAAAGGUCUGACGGAGCAGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCG (((((...........)))))..(((((...((.....))....))).)).((((((((((......((.(((......))).))........)))).)))))).(((((.....))))) ( -36.84) >consensus UGGCCAAAAAAGAGAAGGCCAAAGGCGAAAUGCGUUUAGCUCCAUCGACCGGCUUUACUCAUAAAAAAGUCUGA_GGAGCAGUCCGUAAAAAUUGAGAGAAAGCACGGGAAAAGCUCUCG .(((.......(((((((((...(((((...((.....))....))).)))))))).)))......................(((((.....((....))....)))))....))).... (-23.36 = -23.36 + -0.00)


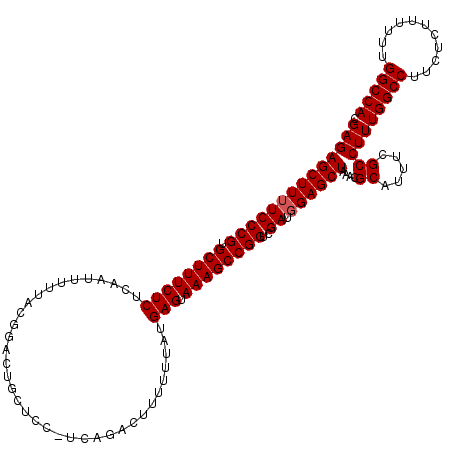
| Location | 21,119,766 – 21,119,886 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.25 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -33.68 |
| Energy contribution | -33.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119766 120 + 23771897 GCUCCGUCAGACUUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCAGGACUGGUGAUGAGUGUGGGUUGCUGUCAUUAUCAGGCUC ((((((((.((((.(((((....)))))..)))))))))))).....((........(((((((...........))))))).((((((((((..((.....))..)))))))))))).. ( -41.80) >DroSec_CAF1 170427 108 + 1 ------------UUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCAGGACUGGCGAUGAGUGUGGGCUGCUGUCAUUAUCAGGCUC ------------..............(((((((.((((((((....((((((((((((((((((...........)))))))...)))))..))))))....))).)))))))).)))). ( -36.50) >DroSim_CAF1 82136 108 + 1 ------------UUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCAGGACUGGCGAUGAGUGUGGGCUGCUGUCAUUAUCAGGCUC ------------..............(((((((.((((((((....((((((((((((((((((...........)))))))...)))))..))))))....))).)))))))).)))). ( -36.50) >DroEre_CAF1 92916 120 + 1 GCUCCCUCAGACCUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCAGGACUAGUGAUGAGUGUGGGCUGCUGUCAUUAUCAGGCUC (((((.((.((((.(((((....)))))..)))))).)))))............((((((((((...........))))))((.(((((((.(((.......)))))))))))))))).. ( -37.30) >DroYak_CAF1 63821 120 + 1 GCUCCGUCAGACCUUUUUAUGAGGAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCAUUUUUGGCCAGGACUAGUGAUGAGUGUGGGCUGCUGUCAUUAUCAGGCUC ((((((((.(((((((((.....)))))..))))))))))))............((((((((((...........))))))((.(((((((.(((.......)))))))))))))))).. ( -40.90) >consensus GCUCC_UCAGACUUUUUUAUGAGUAAAGCCGGUCGAUGGAGCUAAACGCAUUUCGCCUUUGGCCUUCUCUUUUUUGGCCAGGACUGGUGAUGAGUGUGGGCUGCUGUCAUUAUCAGGCUC ..........................(((((((.((((((((....((((((((((((((((((...........)))))))...)))))..))))))....))).)))))))).)))). (-33.68 = -33.84 + 0.16)


| Location | 21,119,886 – 21,120,001 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119886 115 + 23771897 UGGUUGCUCGCCUGCUUUGGCCUAAUUUAUCCCGUCAGUAUGGCAGGAUACCC----GCC-CCCUGAUUGCCUCUUAAUUAUACCCCUUAUAUAUCCACGCUGCACCUGCAGCUUAAAUU .(((.....(((......)))............(((((...(((.((....))----)))-..))))).)))...........................(((((....)))))....... ( -25.40) >DroSec_CAF1 170535 115 + 1 UGGUUGCUCGCCUGCUUUGGCCUAAUUUAUCCCGUCAGUAUCGCAGGAUACCC----GCC-CCCUGAUUGCCUCUUAAUUAUACCCCUUAUAUAUCCGCGCUGCACCUGCAGCUUUAAUU .((((((..(((......)))................(((.(((.(((((...----...-...((((((.....))))))...........)))))))).)))....))))))...... ( -22.55) >DroSim_CAF1 82244 115 + 1 CGGUUGCUCGCCUGCUUUGGCCUAAUUUAUCCCGUCAGUAUGGCAGGAUACCC----GCC-CCCUGAUUGCCUCUUAAUUAUACCCCUUAUAUAUCCGCGCUGCACCUGCAGCUUUAAUU .(((.....(((......)))............(((((...(((.((....))----)))-..))))).)))...........................(((((....)))))....... ( -25.70) >DroEre_CAF1 93036 120 + 1 UGGUCGCUCGCCUGCUUUGGCCUAAUUUAUCCCGUCAGUAUGGCAGGAUACCAUAUUGCCUCCCUCAUUGCCUCCUAAUUAUACCGUUUAUAUAUCCGCGCUGCACCUGCAGCUUUAAUU .((..((..(((......)))............(.((((((((.......)))))))).).........))..))........................(((((....)))))....... ( -25.90) >DroYak_CAF1 63941 115 + 1 UGGUUGCUCGCCUGCUUUGGCCUAAUUUAUCCCGGCAGUGUGGCAGGAUACCC----GCC-ACCUGAUUGCCUCUUAAUUAUACCCCUUAAAUAUCCGCGCUGCACCUGCAGCUUUAAUU .....((..(((......)))............(((((((((((.((....))----)))-))...)))))).........................))(((((....)))))....... ( -32.50) >consensus UGGUUGCUCGCCUGCUUUGGCCUAAUUUAUCCCGUCAGUAUGGCAGGAUACCC____GCC_CCCUGAUUGCCUCUUAAUUAUACCCCUUAUAUAUCCGCGCUGCACCUGCAGCUUUAAUU .((..((..(((......)))......(((((.(((.....))).)))))...................))..))........................(((((....)))))....... (-21.80 = -22.04 + 0.24)



| Location | 21,119,886 – 21,120,001 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -26.44 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21119886 115 - 23771897 AAUUUAAGCUGCAGGUGCAGCGUGGAUAUAUAAGGGGUAUAAUUAAGAGGCAAUCAGGG-GGC----GGGUAUCCUGCCAUACUGACGGGAUAAAUUAGGCCAAAGCAGGCGAGCAACCA .(((((.(((((....))))).)))))......(((((.(((((...(..(..((((..-(((----(((...))))))...))))..)..).))))).)))...((......))..)). ( -34.30) >DroSec_CAF1 170535 115 - 1 AAUUAAAGCUGCAGGUGCAGCGCGGAUAUAUAAGGGGUAUAAUUAAGAGGCAAUCAGGG-GGC----GGGUAUCCUGCGAUACUGACGGGAUAAAUUAGGCCAAAGCAGGCGAGCAACCA .......(((((....))))).....(((((.....)))))...............((.-.((----...(((((((((....)).)))))))......(((......)))..))..)). ( -29.80) >DroSim_CAF1 82244 115 - 1 AAUUAAAGCUGCAGGUGCAGCGCGGAUAUAUAAGGGGUAUAAUUAAGAGGCAAUCAGGG-GGC----GGGUAUCCUGCCAUACUGACGGGAUAAAUUAGGCCAAAGCAGGCGAGCAACCG .......(((((....))))).(((.(((((.....)))))........((..((((..-(((----(((...))))))...)))).............(((......)))..))..))) ( -34.20) >DroEre_CAF1 93036 120 - 1 AAUUAAAGCUGCAGGUGCAGCGCGGAUAUAUAAACGGUAUAAUUAGGAGGCAAUGAGGGAGGCAAUAUGGUAUCCUGCCAUACUGACGGGAUAAAUUAGGCCAAAGCAGGCGAGCGACCA .(((...(((((....)))))...)))........(((......................(((...((..(((((((.(.....).))))))).))...)))...((......)).))). ( -28.80) >DroYak_CAF1 63941 115 - 1 AAUUAAAGCUGCAGGUGCAGCGCGGAUAUUUAAGGGGUAUAAUUAAGAGGCAAUCAGGU-GGC----GGGUAUCCUGCCACACUGCCGGGAUAAAUUAGGCCAAAGCAGGCGAGCAACCA .(((...(((((....)))))...)))......(((((.(((((....((((.....((-(((----(((...))))))))..))))......))))).)))...((......))..)). ( -37.90) >consensus AAUUAAAGCUGCAGGUGCAGCGCGGAUAUAUAAGGGGUAUAAUUAAGAGGCAAUCAGGG_GGC____GGGUAUCCUGCCAUACUGACGGGAUAAAUUAGGCCAAAGCAGGCGAGCAACCA .(((...(((((....)))))...)))........(((......................(((.......(((((((.(.....).)))))))......)))...((......)).))). (-26.44 = -26.64 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:26 2006