| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
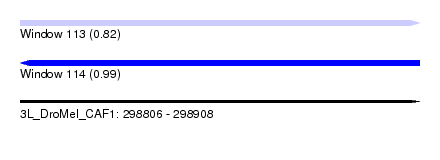
| Location | 298,806 – 298,908 |
| Length | 102 |
| Max. P | 0.992426 |

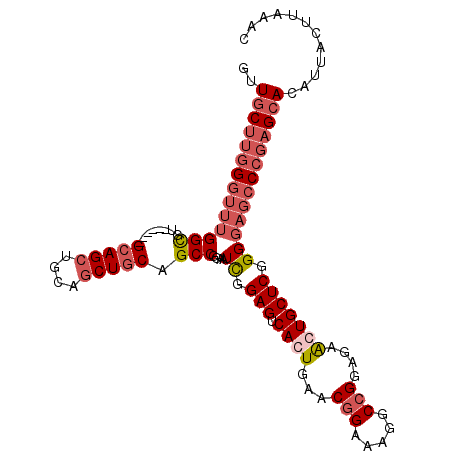
| Location | 298,806 – 298,908 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -22.80 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 298806 102 + 23771897 GUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCAUUCCGUUCAGUGACUCCAAUUCCGGCUGAAGCUGCAGCUGC------ACGCCAAACCCAAGCAAC ((((..((....((((((((...)))))))).......(((((((......))))..........((((((......)))))).------..)))..))..))))... ( -28.80) >DroSec_CAF1 8719 102 + 1 GUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUACGGCCUUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGC------ACACCAAACCCAAGCAAC ((((..((..(((((.(((..((...(((((((...((((.....))))..)))).))).))..)))(((((....))))).))------)))....))..))))... ( -33.40) >DroSim_CAF1 11050 102 + 1 GUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCUUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGC------ACGCCAAACCCAAGCAAC ............((((.(((......(((((((....(((.....)))...)))).))).......(((((((((....)))))------).)))...))).)))).. ( -34.90) >DroYak_CAF1 9691 87 + 1 --------------CUCGGGCUGCCCGAGCAGCUCUCCUGCCACUCCGUUCAGUGACUCCGAUUUCGGCUGCAGUUGCAGCUGCAGCUGCAUGCCA---CU-GGA--- --------------...(((((((....)))))))(((...((((......))))...........((((((((((((....))))))))).))).---..-)))--- ( -37.40) >consensus GUUUAAGUAAUGUGCUCGGGCUCCCCGAGCAGUUCUCCGGCCAUUCCGUUCACUGACUCCGAUUCCGGCUGCAGCUGCAGCUGC______ACGCCAAACCCAAGCAAC ............((((((((...)))))))).......(((......(((....)))........(((((((....))))))).........)))............. (-22.80 = -24.05 + 1.25)

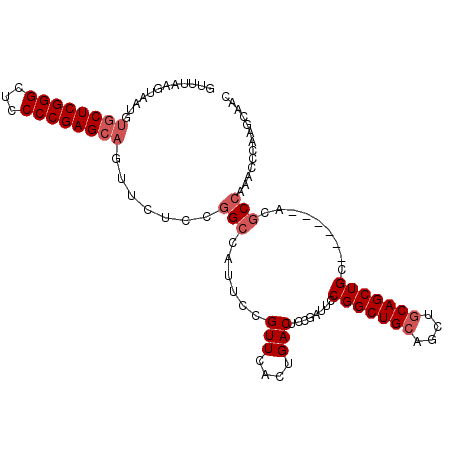
| Location | 298,806 – 298,908 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -26.77 |
| Energy contribution | -29.53 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 298806 102 - 23771897 GUUGCUUGGGUUUGGCGU------GCAGCUGCAGCUUCAGCCGGAAUUGGAGUCACUGAACGGAAUGGCCGGAGAACUGCUCGGGGAGCCCGAGCACAUUACUUAAAC ..(((((((((((..((.------((((((.(.(((....(((...(..(.....)..).)))...))).).))..)))).))..)))))))))))............ ( -36.90) >DroSec_CAF1 8719 102 - 1 GUUGCUUGGGUUUGGUGU------GCAGCUGCAGCUGCAGCCGGAAUCGGAGUCAGUGAACGGAAAGGCCGUAGAACUGCUCGGGGAGCCCGAGCACAUUACUUAAAC ..((((((((((((((.(------(((((....)))))))))....((.(((.((((..((((.....))))...))))))).)))))))))))))............ ( -46.00) >DroSim_CAF1 11050 102 - 1 GUUGCUUGGGUUUGGCGU------GCAGCUGCAGCUGCAGCCGGAAUCGGAGUCAGUGAACGGAAAGGCCGGAGAACUGCUCGGGGAGCCCGAGCACAUUACUUAAAC ..((((((((((((((.(------(((((....)))))))))....((.(((.((((...(((.....)))....))))))).)))))))))))))............ ( -47.60) >DroYak_CAF1 9691 87 - 1 ---UCC-AG---UGGCAUGCAGCUGCAGCUGCAACUGCAGCCGAAAUCGGAGUCACUGAACGGAGUGGCAGGAGAGCUGCUCGGGCAGCCCGAG-------------- ---..(-((---((((.(((((.(((....))).))))).(((....))).)))))))....(((..((......))..)))(((...)))...-------------- ( -38.60) >consensus GUUGCUUGGGUUUGGCGU______GCAGCUGCAGCUGCAGCCGGAAUCGGAGUCACUGAACGGAAAGGCCGGAGAACUGCUCGGGGAGCCCGAGCACAUUACUUAAAC ..((((((((((((((........(((((....))))).)))....((.(((.((((...(((.....)))....))))))).)))))))))))))............ (-26.77 = -29.53 + 2.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:49 2006