
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,114,857 – 21,115,017 |
| Length | 160 |
| Max. P | 0.932664 |

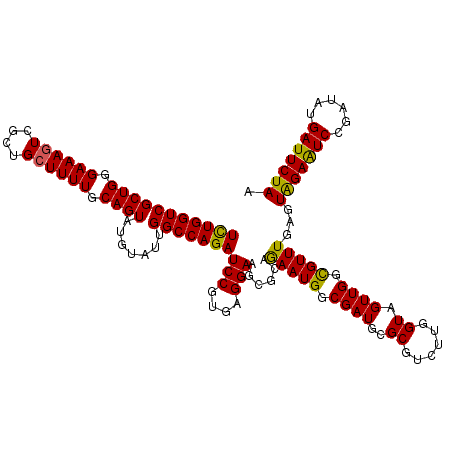
| Location | 21,114,857 – 21,114,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -36.09 |
| Energy contribution | -35.33 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21114857 120 - 23771897 UCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUAAGGAAGCGCAGAAUGGCGAUGCGCGUCUUGGUAGUUGGCGUUUGAGUAGAAUCCGAUAUGAUUCUAA (((((((((((.((((((....)))))).)))).......)))))))(((....))).(((((.........)))))(((.........))).......(((((((......))))))). ( -39.21) >DroSec_CAF1 165457 120 - 1 UCUGGUCGCUGGGAAAGUCACUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUUAGGAAGCGCAGAAUGGCGAUGCGCGUCUUGGUAGUUGGUGUUUGAGUAGAAUCCGAUAUGAUUCUAA (((((((((((.((((((....)))))).)))).......))))))).(..((((((.(((((.........))))).))))))..)............(((((((......))))))). ( -37.51) >DroSim_CAF1 77166 120 - 1 UCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUUAGGAAGCGCAGAAUGGCGAUGCGCGUCUUGGUAGUUGGCGUUUGAGUAGAAUCCGAUAUGAUUCUAA (((((((((((.((((((....)))))).)))).......))))))).(..((((((.(((((.........))))).))))))..)............(((((((......))))))). ( -37.51) >DroEre_CAF1 87982 120 - 1 UCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUGAGGAAUCGCAAAAUGGCGAUGCGCGUCUUGGUAGUUGGCGUUUGAGUGGAGUCCGAGAUGAUUCUAA (((((((((((.((((((....)))))).)))).......)))))))(((....)))(((((......)))))...((((((((.(.((.((......)).)).)))))))))....... ( -45.01) >DroYak_CAF1 58835 120 - 1 UUUGGUCGCUGGGAAAAUCGCUGCUUUUGCAGUACGUAUUGGCCAGAUCCGUGAGGAAGCGCAAAAUGGCGAUGCGCGUCUUGGUAGUUGGCGUUUGAGUGGAAUCCGAGAUGAUUCUAA ((((((((.((.(......(((((....))))).).)).))))))))(((....))).(((((.........)))))(((((((...((.((......)).))..)))))))........ ( -39.40) >consensus UCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUGAGGAAGCGCAGAAUGGCGAUGCGCGUCUUGGUAGUUGGCGUUUGAGUAGAAUCCGAUAUGAUUCUAA (((((((((((.((((((....)))))).)))).......)))))))(((....)))......(((((.((((..((......)).)))).)))))...(((((((......))))))). (-36.09 = -35.33 + -0.76)


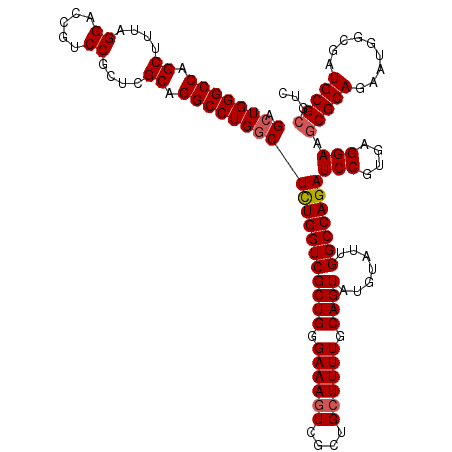
| Location | 21,114,897 – 21,115,017 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -48.61 |
| Consensus MFE | -46.35 |
| Energy contribution | -46.79 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21114897 120 - 23771897 AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGGCUCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUAAGGAAGCGCAGAAUGGCGAUGCGCGUC .((..((((.((....((.....))....)).))))..))(((((((((((.((((((....)))))).)))).......)))))))(((....))).(((((.........)))))... ( -51.51) >DroSec_CAF1 165497 120 - 1 AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGGCUCUGGUCGCUGGGAAAGUCACUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUUAGGAAGCGCAGAAUGGCGAUGCGCGUC .((..((((.((....((.....))....)).))))..))(((((((((((.((((((....)))))).)))).......)))))))(((....))).(((((.........)))))... ( -49.51) >DroSim_CAF1 77206 120 - 1 AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGGCUCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUUAGGAAGCGCAGAAUGGCGAUGCGCGUC .((..((((.((....((.....))....)).))))..))(((((((((((.((((((....)))))).)))).......)))))))(((....))).(((((.........)))))... ( -49.51) >DroEre_CAF1 88022 120 - 1 AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGACUCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUGAGGAAUCGCAAAAUGGCGAUGCGCGUC .((.(((((.((....((.....))....)).)))))...(((((((((((.((((((....)))))).)))).......)))))))(((....)))(((((......)))))..))... ( -45.31) >DroYak_CAF1 58875 120 - 1 AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGGCUUUGGUCGCUGGGAAAAUCGCUGCUUUUGCAGUACGUAUUGGCCAGAUCCGUGAGGAAGCGCAAAAUGGCGAUGCGCGUC .((..((((.((....((.....))....)).))))..))((((((((.((.(......(((((....))))).).)).))))))))(((....))).(((((.........)))))... ( -47.20) >consensus AGCUGGGCCAGCUUUAGGACCGUCCGCUCGCAGGCCUGGCUCUGGUCGCUGGGAAAGUCGCUGCUUUUGCAGUAUGUAUUGGCCAGAUCCGUGAGGAAGCGCAGAAUGGCGAUGCGCGUC .((((((((.((....((.....))....)).))))))))(((((((((((.((((((....)))))).)))).......)))))))(((....))).(((((.........)))))... (-46.35 = -46.79 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:04 2006