
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,389,970 – 2,390,078 |
| Length | 108 |
| Max. P | 0.838715 |

| Location | 2,389,970 – 2,390,078 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -49.55 |
| Consensus MFE | -40.51 |
| Energy contribution | -40.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
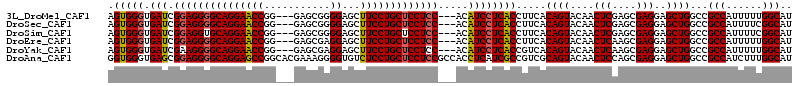
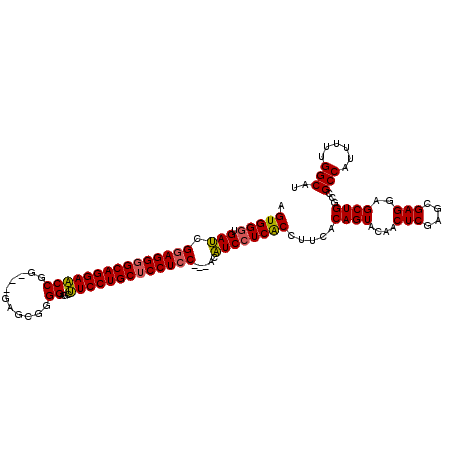
>3L_DroMel_CAF1 2389970 108 + 23771897 AGUGGGUGAUCGGAGGGGCAGGAACCGG---GAGCGGGGAGCUUCCUGCUCCUCC---ACAUCCUCACCUUCACAGUACAACUCGAGCGAGGAGCUGGCCGCCAUUUUUGGCAU .(((((.(((.((((((((((((((((.---...))).....)))))))))))))---..)))))))).....((((....(((....)))..))))...((((....)))).. ( -49.40) >DroSec_CAF1 16441 108 + 1 AGUGGGUGAUCGGAGGGGCAGGAACCGG---GAGCGGGGAGCUUCCUGCUCCUCC---ACAUCCUCACCUUCACAGUACAACUCGAGCGAGGAGCUGGCCGCCAUUUUCGGCAU .(((((.(((.((((((((((((((((.---...))).....)))))))))))))---..)))))))).....((((....(((....)))..))))((((.......)))).. ( -48.50) >DroSim_CAF1 16530 108 + 1 AGUGGGUGAUCGGAGGUGCAGGAACCGG---GAGCGGGGAGCUUCCUGCUCCUCC---ACAUCCUCACCUUCACAGUACAACUCGAGCGAGGAGCUGGCCGCCAUUUUCGGCAU ....(((....(((((((.((((...((---((((((((....))))))))))..---...))))))))))).((((....(((....)))..)))))))(((......))).. ( -47.60) >DroEre_CAF1 16499 108 + 1 AGUGGGUGAUCGGAGGGGCAGGAACCGG---GAGCGAGGAGCUUCCUGCUCCUCC---ACAUCCUCACCUUCACAGUACAACUCAAGCGAGGAGCUGGCCGCCAUUUUUGGCAU .(((((.(((.(((((((((((((((..---......))...)))))))))))))---..)))))))).....((((....(((....)))..))))...((((....)))).. ( -46.60) >DroYak_CAF1 16777 108 + 1 AGUGGGUGAUCGAAGGGGCAGGAACCGG---GAGCGAGGAGCUUCCUGCUCCUCC---ACAUCCUCACCGUCACAGUACAACUCAAGCGAGGAGCUGGCCGCCAUUUUUGGCAU .(((((.(((.(.(((((((((((((..---......))...))))))))))).)---..)))))))).....((((....(((....)))..))))...((((....)))).. ( -39.80) >DroAna_CAF1 16069 114 + 1 GGUGGGUGAGCGGAGGGGCAGGAGCCGGCACGAAAGGGGUGUCUCCUGCUCCUCCGCCACCUCAUCGCCGUCGCAGUACAACUCCAGCGAGGAGCUGGCCGCCAUCUUUGGCAU ((((((((.(((((((((((((((.((.(.(....).).)).))))))))))))))))))).))))((....)).........(((((.....)))))..((((....)))).. ( -65.40) >consensus AGUGGGUGAUCGGAGGGGCAGGAACCGG___GAGCGGGGAGCUUCCUGCUCCUCC___ACAUCCUCACCUUCACAGUACAACUCGAGCGAGGAGCUGGCCGCCAUUUUUGGCAU .(((((.(((.(((((((((((((((...........))...))))))))))))).....)))))))).....((((....(((....)))..))))...(((......))).. (-40.51 = -40.32 + -0.19)
| Location | 2,389,970 – 2,390,078 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -45.63 |
| Consensus MFE | -38.32 |
| Energy contribution | -37.71 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
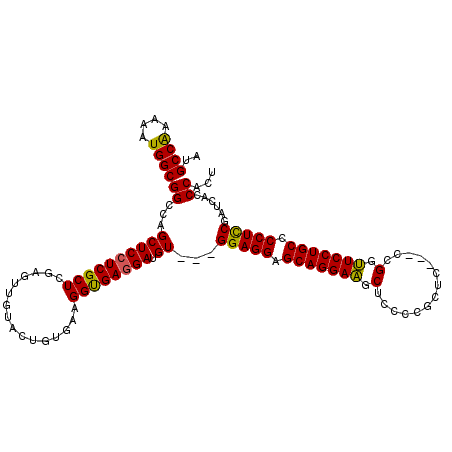
>3L_DroMel_CAF1 2389970 108 - 23771897 AUGCCAAAAAUGGCGGCCAGCUCCUCGCUCGAGUUGUACUGUGAAGGUGAGGAUGU---GGAGGAGCAGGAAGCUCCCCGCUC---CCGGUUCCUGCCCCUCCGAUCACCCACU ..((((....))))(..((((((.......))))))..).(((..(((((.....(---(((((.(((((((.....(((...---.)))))))))).)))))).)))))))). ( -44.40) >DroSec_CAF1 16441 108 - 1 AUGCCGAAAAUGGCGGCCAGCUCCUCGCUCGAGUUGUACUGUGAAGGUGAGGAUGU---GGAGGAGCAGGAAGCUCCCCGCUC---CCGGUUCCUGCCCCUCCGAUCACCCACU ..(((......)))(..((((((.......))))))..).(((..(((((.....(---(((((.(((((((.....(((...---.)))))))))).)))))).)))))))). ( -43.30) >DroSim_CAF1 16530 108 - 1 AUGCCGAAAAUGGCGGCCAGCUCCUCGCUCGAGUUGUACUGUGAAGGUGAGGAUGU---GGAGGAGCAGGAAGCUCCCCGCUC---CCGGUUCCUGCACCUCCGAUCACCCACU ..(((......)))(..((((((.......))))))..).(((..((((((((.((---(.((((((.((.(((.....))).---)).)))))).))).)))..)))))))). ( -44.40) >DroEre_CAF1 16499 108 - 1 AUGCCAAAAAUGGCGGCCAGCUCCUCGCUUGAGUUGUACUGUGAAGGUGAGGAUGU---GGAGGAGCAGGAAGCUCCUCGCUC---CCGGUUCCUGCCCCUCCGAUCACCCACU ..((((....))))(..((((((.......))))))..).(((..(((((.....(---(((((.(((((((.(.........---..).))))))).)))))).)))))))). ( -43.80) >DroYak_CAF1 16777 108 - 1 AUGCCAAAAAUGGCGGCCAGCUCCUCGCUUGAGUUGUACUGUGACGGUGAGGAUGU---GGAGGAGCAGGAAGCUCCUCGCUC---CCGGUUCCUGCCCCUUCGAUCACCCACU ..((((....))))(..((((((.......))))))..).(((..(((((.....(---(((((.(((((((.(.........---..).))))))).)))))).)))))))). ( -40.90) >DroAna_CAF1 16069 114 - 1 AUGCCAAAGAUGGCGGCCAGCUCCUCGCUGGAGUUGUACUGCGACGGCGAUGAGGUGGCGGAGGAGCAGGAGACACCCCUUUCGUGCCGGCUCCUGCCCCUCCGCUCACCCACC .(((((....)))))((((.(((.((((((..((......))..)))))).))).))))(((((.(((((((.(((.......)))....))))))).)))))........... ( -57.00) >consensus AUGCCAAAAAUGGCGGCCAGCUCCUCGCUCGAGUUGUACUGUGAAGGUGAGGAUGU___GGAGGAGCAGGAAGCUCCCCGCUC___CCGGUUCCUGCCCCUCCGAUCACCCACU ..((((....))))((...((((((((((................)))))))).))...(((((.(((((((.(..............).))))))).)))))......))... (-38.32 = -37.71 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:29 2006