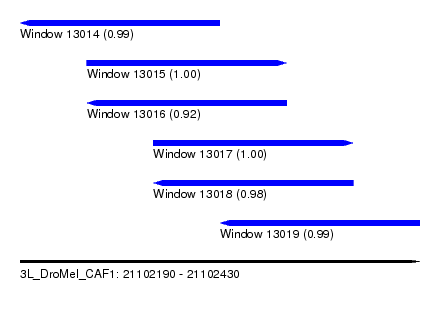
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,102,190 – 21,102,430 |
| Length | 240 |
| Max. P | 0.999958 |

| Location | 21,102,190 – 21,102,310 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.26 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102190 120 - 23771897 GGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAGUGAAAAGUAUACACCAAUUUGGUAGGGUAACCGGUCAAAA ((((..(((....((((.((((.(((((.(((.....)))...............(((.....))).))))).))))))))....)))....(((.....))).((....)))))).... ( -35.40) >DroSec_CAF1 152452 120 - 1 GGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUGUUAAUCCGGGCUGGAGGAUGCUGCCACAGUGAAAAUUACACACCAAUUUGGCAGUGUAACCGGUCAAAA ((((.((......(((.(((((.(((((.(((.....)))((((.....))))..(((.....))).))))).)))))))).....((((((.((.....))..)))))))))))).... ( -39.70) >DroSim_CAF1 59995 120 - 1 GGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAGUGAAAAGUACACACCAAUUUGGCAGUGUAACCGGUCAAAA (((((.(.........).))))).....(((...(((((........)))))....)))((((((...((((((((((((((.........)))...)))))))))))..)))))).... ( -37.60) >DroEre_CAF1 74793 119 - 1 GGCCAGGCUCAAAACUUGUGGCUGCAUCGGCCACAGAGCCAACAAUUGCUUUUAAUCCC-GCUGGAGGAUACUGCCAAAGUAAAAAGUACACACAAAUUUGGCAGGGCAGCCGGUCAAUA ((((.(((((......((((((((...))))))))))))).....(((((....(((((-....).)))).((((((((..................)))))))))))))..)))).... ( -42.77) >DroYak_CAF1 46875 120 - 1 GGCCGGGCUCAAAACUUGUGGCUGCAUCGGCCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUACUGCCAAAGUGAAAACUACACACCAAUUUGGCAGGGUAGCCGGUCUAAA ((((((.(((.....(((((((((...)))))))))((((...................)))).)))....(((((((((((.........)))...)))))))).....)))))).... ( -41.91) >consensus GGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAGUGAAAAGUACACACCAAUUUGGCAGGGUAACCGGUCAAAA ((((.((((......(((((.(((...))).))))))))).....((((.....((((........)))).(((((((((((.........)))...)))))))).))))..)))).... (-32.58 = -32.26 + -0.32)


| Location | 21,102,230 – 21,102,350 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -45.01 |
| Consensus MFE | -38.42 |
| Energy contribution | -39.86 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102230 120 + 23771897 CUUUGGCAGCAUCCUCCAGCCGGGAUUAAAAGCAAUUGUUGGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUCUGUGCCGCUGCCAAGAGGCAA .(((((((((........((((((....(((((........)))))...))))..)).(.((((..(.((((((((((......))))))).))).)..)))).))))))))))...... ( -45.40) >DroSec_CAF1 152492 120 + 1 CUGUGGCAGCAUCCUCCAGCCCGGAUUAACAGCAAUUGUUGGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAA ..((((((.((......((((((((((....(((.(((..(((.....))))))))).((((...((.....)).))))..))))))))))........))))))))((((....)))). ( -44.64) >DroSim_CAF1 60035 120 + 1 CUUUGGCAGCAUCCUCCAGCCGGGAUUAAAAGCAAUUGUUGGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAA .(((((((((........((((((....(((((........)))))...))))..)).(.(((((((.((((((((((......))))))).))).))))))).))))))))))...... ( -48.50) >DroEre_CAF1 74833 119 + 1 CUUUGGCAGUAUCCUCCAGC-GGGAUUAAAAGCAAUUGUUGGCUCUGUGGCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCUAGGCUGCAAUAUUUGAGCCGCCGCCAAGAGCCAA .....((...(((((.....-))))).....)).....((((((((.((((....((.((..(((((.((((((((((......))))))).))).))))).)).)).)))))))))))) ( -42.30) >DroYak_CAF1 46915 120 + 1 CUUUGGCAGUAUCCUCCAGCCGGGAUUAAAAGCAAUUGUUGGCUUUGUGGCCGAUGCAGCCACAAGUUUUGAGCCCGGCCAAAUCUAGGCUGUAAUAUUUGUGCCGCUGCCAAGAGCGAA .(((((((((........((((((.((((((((........))((((((((.......)))))))))))))).))))))........(((............))))))))))))...... ( -44.20) >consensus CUUUGGCAGCAUCCUCCAGCCGGGAUUAAAAGCAAUUGUUGGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAA .(((((((((........((((((....(((((........)))))...))))..)).(.(((((((.((((((((((......))))))).))).))))))).))))))))))...... (-38.42 = -39.86 + 1.44)



| Location | 21,102,230 – 21,102,350 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -33.51 |
| Energy contribution | -34.15 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102230 120 - 23771897 UUGCCUCUUGGCAGCGGCACAGAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAG .......(((((((((.(((((.......((((.((......)).))))......))))).((.(((((((...(((((........)))))....))))).)).))..))))))))).. ( -40.42) >DroSec_CAF1 152492 120 - 1 UUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUGUUAAUCCGGGCUGGAGGAUGCUGCCACAG .((((....))))((((((.........(((((((((((......((((......(((((.(((...))).)))))))))((((.....))))))))))))))).....))))))..... ( -45.44) >DroSim_CAF1 60035 120 - 1 UUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAG .......(((((((((.(((((.......((((.((......)).))))......))))).((.(((((((...(((((........)))))....))))).)).))..))))))))).. ( -40.72) >DroEre_CAF1 74833 119 - 1 UUGGCUCUUGGCGGCGGCUCAAAUAUUGCAGCCUAGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGCCACAGAGCCAACAAUUGCUUUUAAUCCC-GCUGGAGGAUACUGCCAAAG .......(((((((.(((((.....(((.(((((.(......).))))))))....((((((((...)))))))))))))..............(((((-....).)))).))))))).. ( -42.50) >DroYak_CAF1 46915 120 - 1 UUCGCUCUUGGCAGCGGCACAAAUAUUACAGCCUAGAUUUGGCCGGGCUCAAAACUUGUGGCUGCAUCGGCCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUACUGCCAAAG .......(((((((.(((............)))....((..((((((..........(((((((...)))))))(((((........)))))....))))))..)).....))))))).. ( -43.50) >consensus UUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCCAACAAUUGCUUUUAAUCCCGGCUGGAGGAUGCUGCCAAAG .......(((((((((............(((((.(((((.(((..((((......(((((.(((...))).))))))))).......)))...))))).))))).....))))))))).. (-33.51 = -34.15 + 0.64)



| Location | 21,102,270 – 21,102,390 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -40.99 |
| Consensus MFE | -38.50 |
| Energy contribution | -38.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102270 120 + 23771897 GGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUCUGUGCCGCUGCCAAGAGGCAAGUAAAAAGUGGCAGCAACCAGGACAAUUAGCACAGCCAAA ((((..((((((..(((.(((((..(((...(((((((......)))))))..))).....((((.((....)).))))........))))).)))....)).......))))))))... ( -42.61) >DroSec_CAF1 152532 120 + 1 GGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAAGUAAAAAGUGGCAGCAACCAGGACAAUUAGCACAGCCAAA ((((.(((((((...((((((((((((.((((((((((......))))))).))).)))))))..)))))...).)))..........(((......))).........))).))))... ( -44.00) >DroSim_CAF1 60075 120 + 1 GGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAAGUAAAAAGUGGCAGCAACCAGGACAAUUAGCACAGCCAAA ((((.(((((((...((((((((((((.((((((((((......))))))).))).)))))))..)))))...).)))..........(((......))).........))).))))... ( -44.00) >DroEre_CAF1 74872 120 + 1 GGCUCUGUGGCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCUAGGCUGCAAUAUUUGAGCCGCCGCCAAGAGCCAAGUAAAAAGCAACAGCAACUAGGACAAUUAGCACAGCCAAA ((((((.((((....((.((..(((((.((((((((((......))))))).))).))))).)).)).)))))))))).........((....))..((((.....)))).......... ( -37.50) >DroYak_CAF1 46955 120 + 1 GGCUUUGUGGCCGAUGCAGCCACAAGUUUUGAGCCCGGCCAAAUCUAGGCUGUAAUAUUUGUGCCGCUGCCAAGAGCGAAGUAGAAAGUAACAGCAACUAGGACAAUUAGCACAGCCAAA ((((.((((((((..((...............)).)))))...((((((((((....(((.(((((((......))))..))).)))...)))))..))))).......))).))))... ( -36.86) >consensus GGCUUUGUGCCCGAUGCAGCCACAAGUUUUGAGCCUGGCCAAAUCCGGGCUAUAAUAUUUGUGCCGCUGCCAAGAGGCAAGUAAAAAGUGGCAGCAACCAGGACAAUUAGCACAGCCAAA ((((.(((((((...((((((((((((.((((((((((......))))))).))).)))))))..)))))...).))).........((....))..............))).))))... (-38.50 = -38.34 + -0.16)



| Location | 21,102,270 – 21,102,390 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -34.91 |
| Energy contribution | -33.59 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102270 120 - 23771897 UUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAGAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCC ...((((.((.........(((((((((((((................)))))))))).)))........(((((.((.((((((.(.........).)))))).))))))).)).)))) ( -41.69) >DroSec_CAF1 152532 120 - 1 UUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCC ...(((((((((...(((((..((((((((((................))))))))))((((.......((((.((......)).))))......))))))..)))...)))))..)))) ( -39.21) >DroSim_CAF1 60075 120 - 1 UUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCC ...(((((((((...(((((..((((((((((................))))))))))((((.......((((.((......)).))))......))))))..)))...)))))..)))) ( -39.21) >DroEre_CAF1 74872 120 - 1 UUUGGCUGUGCUAAUUGUCCUAGUUGCUGUUGCUUUUUACUUGGCUCUUGGCGGCGGCUCAAAUAUUGCAGCCUAGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGCCACAGAGCC ..((((((..(((.........(((((((..(((........)))...)))))))((((((.....)).)))))))...))))))(((((......((((((((...))))))))))))) ( -42.80) >DroYak_CAF1 46955 120 - 1 UUUGGCUGUGCUAAUUGUCCUAGUUGCUGUUACUUUCUACUUCGCUCUUGGCAGCGGCACAAAUAUUACAGCCUAGAUUUGGCCGGGCUCAAAACUUGUGGCUGCAUCGGCCACAAAGCC ...(((((((.((.((((....((((((((((................)))))))))))))).)).)))))))............((((......(((((((((...))))))))))))) ( -38.69) >consensus UUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUUGGCCAGGCUCAAAACUUGUGGCUGCAUCGGGCACAAAGCC ...(((((((.((.((((....((((((((((................)))))))))))))).)).)))))))............((((......(((((.(((...))).))))))))) (-34.91 = -33.59 + -1.32)



| Location | 21,102,310 – 21,102,430 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -35.27 |
| Energy contribution | -34.27 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21102310 120 - 23771897 UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCAGUUUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAGAUAUUAUAGCCCGGAUUU .....((((....((((..(....)...)))).))))((((..(((((((.....((((.((((((((((((................)))))))))).)))))).)))))))..)))). ( -38.59) >DroSec_CAF1 152572 120 - 1 UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCACUUUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUU .((((........((((..(....)...))))...))))((..(((((((.((.((((....((((((((((................)))))))))))))).)).)))))))..))... ( -36.89) >DroSim_CAF1 60115 120 - 1 UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCACUUUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUU .((((........((((..(....)...))))...))))((..(((((((.((.((((....((((((((((................)))))))))))))).)).)))))))..))... ( -36.89) >DroEre_CAF1 74912 120 - 1 UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCAGUUUUGGCUGUGCUAAUUGUCCUAGUUGCUGUUGCUUUUUACUUGGCUCUUGGCGGCGGCUCAAAUAUUGCAGCCUAGAUUU .....((((....((((..(....)...)))).))))(((((.((((((..((.(((....(((((((((((((........)))....))))))))))))).))..)))))).))))). ( -35.20) >DroYak_CAF1 46995 120 - 1 UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCAGUUUUGGCUGUGCUAAUUGUCCUAGUUGCUGUUACUUUCUACUUCGCUCUUGGCAGCGGCACAAAUAUUACAGCCUAGAUUU .....((((....((((..(....)...)))).))))(((((.(((((((.((.((((....((((((((((................)))))))))))))).)).))))))).))))). ( -34.49) >consensus UGUGCGUUGUCUUGCCAACGGAAACUUAUGGCCCAGCAGUUUUGGCUGUGCUAAUUGUCCUGGUUGCUGCCACUUUUUACUUGCCUCUUGGCAGCGGCACAAAUAUUAUAGCCCGGAUUU .....((((....((((..(....)...)))).))))(((((.(((((((.((.((((....((((((((((................)))))))))))))).)).))))))).))))). (-35.27 = -34.27 + -1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:39 2006