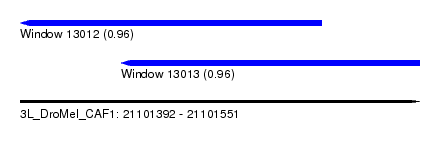
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,101,392 – 21,101,551 |
| Length | 159 |
| Max. P | 0.963506 |

| Location | 21,101,392 – 21,101,512 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -28.59 |
| Energy contribution | -28.99 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
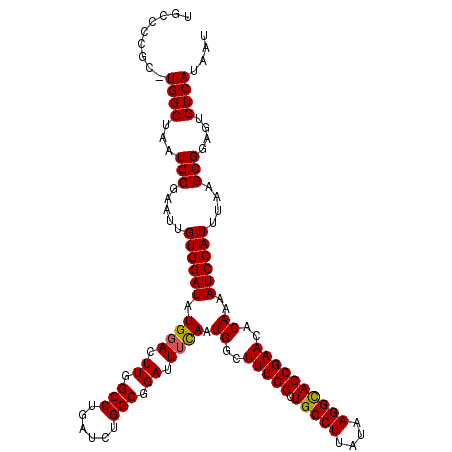
>3L_DroMel_CAF1 21101392 120 - 23771897 CCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAAUGACUCACUACUGCUCGCCAGCCAAG .((...(((((((((((...((..(((((.(((((....))))))))))..)))))))).....(((((((((((((................)))))..))).))))).)))))...)) ( -37.49) >DroSec_CAF1 151641 117 - 1 CCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAGUGACUCAC---UGGUCGCCAGCCAAG .((...(((((((((((...((..(((((.(((((....))))))))))..)))))))).......(((.(.(((((................))))).).)---))...)))))...)) ( -34.19) >DroSim_CAF1 59185 117 - 1 CCUGAUCUGGCGGACUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAAUGACUCAC---UGCUCGCCAGCCAAG .((...((((((((.((...((..(((((.(((((....))))))))))..)))).))).....(((((.(.(((((................))))).).)---)))).)))))...)) ( -34.09) >DroEre_CAF1 73940 117 - 1 CCUGAUCAGGCGGAUUUUAAUGGCUUUGGCUGCCUUAUAAGGUACCGAAAACAAUAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAAUGACUCAC---UGUUCGCCAGCCAAG ........(((((((.....((..(((((.(((((....))))))))))..))..)))).....(((((.(.(((((................))))).).)---)))).)))....... ( -29.09) >DroYak_CAF1 45939 117 - 1 CCUGAUCUGGCGGAUUGUAAUGGCUUUGGCUGCCUUAUAAGGUACCGAAAACAAAAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAAUGACUCAC---UGUUCGCCAGCCAAG .((...((((((((((....((..(((((.(((((....))))))))))..)).))))).....(((((.(.(((((................))))).).)---)))).)))))...)) ( -33.89) >consensus CCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAUAUAAAACCUCGGAAUGACUCAC___UGCUCGCCAGCCAAG .((...(((((((((((...((..(((((.(((((....))))))))))..))))))))...........(.(((((................))))).)..........)))))...)) (-28.59 = -28.99 + 0.40)

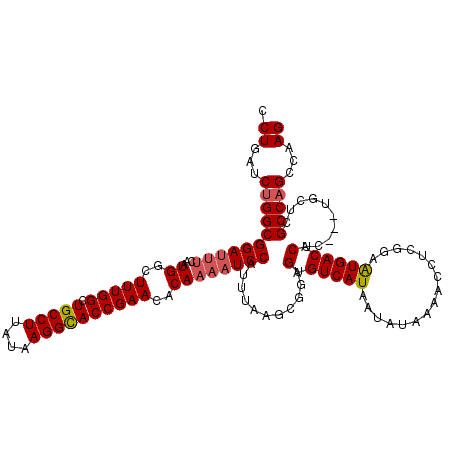
| Location | 21,101,432 – 21,101,551 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -34.56 |
| Energy contribution | -34.28 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21101432 119 - 23771897 UGCCCCGC-UGGCUAAUGCGAAUUGUGGAUAUGGACUUGGCCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAU .((.((((-(.((....)).....((((((.((((.((.(((......))).)).)))).((..(((((.(((((....))))))))))..))..))))))...))))).))........ ( -39.40) >DroSec_CAF1 151678 119 - 1 UGCCCCGC-UGGCUAAUGCGAAUUGUGGAUAUGGACUUGGCCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAU .((.((((-(.((....)).....((((((.((((.((.(((......))).)).)))).((..(((((.(((((....))))))))))..))..))))))...))))).))........ ( -39.40) >DroSim_CAF1 59222 119 - 1 UGCCCCGC-UGGCUAAUGCGAAUUGUGGAUAUGGACUUGGCCUGAUCUGGCGGACUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAU .((.((((-(.((....)).....((((((.((((.((.(((......))).)).)))).((..(((((.(((((....))))))))))..))..))))))...))))).))........ ( -39.40) >DroEre_CAF1 73977 119 - 1 UGCCCCAC-UGGCUAAUGCGAAUUGUGGAUAUGGACUUGGCCUGAUCAGGCGGAUUUUAAUGGCUUUGGCUGCCUUAUAAGGUACCGAAAACAAUAUCCAUUUAAGCGGAGUGUCAUAAU ........-((((...(((.....(((((((((((.((.((((....)))).)).)))..((..(((((.(((((....))))))))))..))))))))))....)))....)))).... ( -35.10) >DroYak_CAF1 45976 120 - 1 UACCCCGUGUGGCUAAUGCGAGUUGUGGAUAUGGACUUGGCCUGAUCUGGCGGAUUGUAAUGGCUUUGGCUGCCUUAUAAGGUACCGAAAACAAAAUCCAUUUAAGCGGAGUGUCAUAAU .......((((((...(((.....((((((.((.(.((.(((......))).)).).)).((..(((((.(((((....))))))))))..))..))))))....)))....)))))).. ( -34.10) >consensus UGCCCCGC_UGGCUAAUGCGAAUUGUGGAUAUGGACUUGGCCUGAUCUGGCGGAUUUCAAUGGCUUUGGCUGCCUUAUAAGGCACCGAACACAAAAUCCAUUUAAGCGGAGUGUCAUAAU .........((((...(((.....((((((.((((.((.(((......))).)).)))).((..(((((.(((((....))))))))))..))..))))))....)))....)))).... (-34.56 = -34.28 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:33 2006