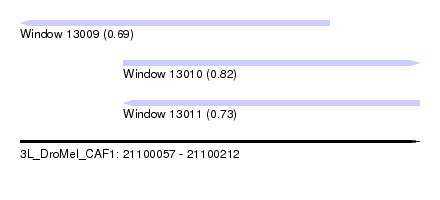
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,100,057 – 21,100,212 |
| Length | 155 |
| Max. P | 0.818784 |

| Location | 21,100,057 – 21,100,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.41 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -18.16 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21100057 120 - 23771897 CUAUAAAUGUGGGCCGAAAAAUGGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAAUGAGUUAAUUUUUUGAUUAAAUCUCGCGCUACC ...........((((((...((((....(((((((..........)))))))....))))..((((..((.((((((...((.....))..)))))).))..))))....))).)))... ( -23.80) >DroSec_CAF1 150379 120 - 1 CUAUAAAAAUUGGCCGAAAAAUGAGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAAUGAGUUAAUUUUUUGAUUAAAUCUCGCGCUACC ..........(((((((...((((....(((((((..........)))))))....))))..((((..((.((((((...((.....))..)))))).))..))))....))).)))).. ( -25.30) >DroSim_CAF1 57851 120 - 1 CUAUAAAAAUUGGCCGAAAAAUGAGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAAUGAGUUAAUUUUUUGAUUAAAUCUCGCGCUACC ..........(((((((...((((....(((((((..........)))))))....))))..((((..((.((((((...((.....))..)))))).))..))))....))).)))).. ( -25.30) >DroEre_CAF1 72550 119 - 1 CUAUAUAGAUUGGCCGAAAAAAUGGAAAGCGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAACGAGUUAAUU-UUUGAUUAAAUCUCCCGCUACC ..........((((.......((((...(((((((..........)))))))....))))..........((((((((((...((.....))...))-))))))))........)))).. ( -20.30) >DroYak_CAF1 44527 119 - 1 CCAUAUAAAUUGGGCGAAAAAAUGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAAUGAGUUAAUU-UUUGAUAAAAUCUCCUGCUACC (((.......)))..........((((.(((((((..........)))))))...((((....((((((...(((..........)))...))))))-..))))......))))...... ( -19.70) >consensus CUAUAAAAAUUGGCCGAAAAAUGGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAACACACAAUGAGUUAAUUUUUUGAUUAAAUCUCGCGCUACC ..........(((((((...........(((((((..........)))))))..........((((..((.((((((...((.....))..)))))).))..))))....))).)))).. (-18.16 = -19.20 + 1.04)


| Location | 21,100,097 – 21,100,212 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -19.98 |
| Energy contribution | -21.02 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21100097 115 + 23771897 UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGUUUCCCCAUUUUUCGGCCCACAUUUAUAGGUCCG----ACUCAUUCG-GGGCGUGUUUUAUCAUAGCCG ..................((((((....(((((((((......)))))))))..((((.....((((.((.........)).)))----).......)-))).........))))))... ( -29.30) >DroSec_CAF1 150419 115 + 1 UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGUUUCCUCAUUUUUCGGCCAAUUUUUAUAGGUCCG----ACUUAUUCG-GGGCGUGUUUUAUCAUAGCCG ....................((((....(((((((((......)))))))))....))))....((((...........(.((((----(.....)))-)).)(((......))).)))) ( -29.10) >DroSim_CAF1 57891 115 + 1 UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGUUUCCUCAUUUUUCGGCCAAUUUUUAUAGGUCCG----ACUCAUUCG-GGGCGUGUUUUAUCAUAGCCG ....................((((....(((((((((......)))))))))....))))....((((...........(.((((----(.....)))-)).)(((......))).)))) ( -29.10) >DroEre_CAF1 72589 120 + 1 UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGCUUUCCAUUUUUUCGGCCAAUCUAUAUAGGUACAUGGUACAUAUUCAUGGGCGUGUUUUAUCAUAGCCG ..................((((((.....((((((((......))))))))...............(((.((((....)))).(((((.......))))))))........))))))... ( -25.80) >DroYak_CAF1 44566 114 + 1 UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGUUUCCCAUUUUUUCGCCCAAUUUAUAUGGGCCCG----ACUCAUU--UAGGCGGAUUUUUUCAUAGCCG ....................((((....(((((((((......))))))))).............(((((.......)))))...----..)))).--..(((.((.....))...))). ( -28.20) >consensus UUUUUAAUUAUUCAAUUAUUAUGACUUUACGGCUGUUUGCCUUGGCAGCCGUUUCCCCAUUUUUCGGCCAAUUUUUAUAGGUCCG____ACUCAUUCG_GGGCGUGUUUUAUCAUAGCCG ..................((((((....(((((((((......)))))))))..............(((.......((((((.......)))))).....)))........))))))... (-19.98 = -21.02 + 1.04)



| Location | 21,100,097 – 21,100,212 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21100097 115 - 23771897 CGGCUAUGAUAAAACACGCCC-CGAAUGAGU----CGGACCUAUAAAUGUGGGCCGAAAAAUGGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA ....((((((........(((-((......(----(((.(((((....)))))))))....)))))..(((((((..........)))))))...))))))................... ( -34.40) >DroSec_CAF1 150419 115 - 1 CGGCUAUGAUAAAACACGCCC-CGAAUAAGU----CGGACCUAUAAAAAUUGGCCGAAAAAUGAGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA ....((((((..........(-(..((...(----(((.((..........))))))...))..))..(((((((..........)))))))...))))))................... ( -25.20) >DroSim_CAF1 57891 115 - 1 CGGCUAUGAUAAAACACGCCC-CGAAUGAGU----CGGACCUAUAAAAAUUGGCCGAAAAAUGAGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA ....((((((..........(-(..((...(----(((.((..........))))))...))..))..(((((((..........)))))))...))))))................... ( -26.30) >DroEre_CAF1 72589 120 - 1 CGGCUAUGAUAAAACACGCCCAUGAAUAUGUACCAUGUACCUAUAUAGAUUGGCCGAAAAAAUGGAAAGCGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA ((((((...........(..((((.........))))..)..........)))))).....((((...(((((((..........)))))))....)))).................... ( -22.50) >DroYak_CAF1 44566 114 - 1 CGGCUAUGAAAAAAUCCGCCUA--AAUGAGU----CGGGCCCAUAUAAAUUGGGCGAAAAAAUGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA .((((..........(((.((.--....)).----)))(((((.......))))).............(((((((..........)))))))..))))...................... ( -27.00) >consensus CGGCUAUGAUAAAACACGCCC_CGAAUGAGU____CGGACCUAUAAAAAUUGGCCGAAAAAUGGGGAAACGGCUGCCAAGGCAAACAGCCGUAAAGUCAUAAUAAUUGAAUAAUUAAAAA (((((............(((...............(((.((..........)))))........(....))))(((....)))...))))).....(((.......)))........... (-18.34 = -18.58 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:31 2006