
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
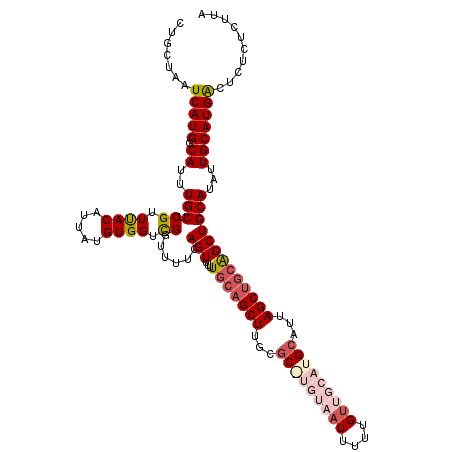
| Location | 21,098,646 – 21,098,806 |
| Length | 160 |
| Max. P | 0.989145 |

| Location | 21,098,646 – 21,098,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.96 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -25.26 |
| Energy contribution | -27.54 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21098646 120 + 23771897 CUGCUAAUCAUGACAUUUGCUCGUUUACAUUAUGUGGUCGGUUUUCAGUUUUUGCAGCUUGCGCAUGUAAUUUUUGAUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGGCUCUCUCUUA .......(((((.((..((((((.((((.....)))).))).....(((...(((((((...(((((((........)))))))...)))))))))))))...))))))).......... ( -32.50) >DroSec_CAF1 148832 119 + 1 CUGCUAAUCAUGGCAUUUGCUCGUUUACAUUAUGUGGUCGGUUUUCAGUUUUU-CAGCUUGCGCAUGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUA .......(((((.((..((((((.((((.....)))).))).....((((...-(((((...(((((((((....)))))))))...))))).)))))))...))))))).......... ( -32.10) >DroSim_CAF1 56361 120 + 1 CUGCUAAUCAUGGCAUUUGCUCGUUUACAUUAUGUGGUCGGUUUUCAGUUUUUGCAGCUUGCGCGUGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUA .......(((((.((..((((((.((((.....)))).))).....(((...(((((((...(((((((((....)))))))))...)))))))))))))...))))))).......... ( -35.80) >DroEre_CAF1 71196 115 + 1 CUGCUAAUCAUGGCAUUUGCUCGUUUACAUUAUGUGGUCGGUUUUCAGUUUUUGCAGCUUGUGC-----AUUUUUGUUGCAUGCAUCAGCCACGGCUGCAUAUUGCAUGACUUUCCCUUG .......(((((((....)).......((.((((..((((.......((....)).(((.((((-----((.........)))))).)))..))))..)))).))))))).......... ( -28.20) >DroYak_CAF1 43072 115 + 1 UUGCUAAUCAUGGCAUUUGCUCGUUCACAUUAUGUGGUUGCAGCUCAGUUUUUGCAGCUUGUUC-----AUUUUUGUUGCAUGCAUCAGCUGCAGCUGCAUAUUGCAUGACUUUCCCUUA .......(((((((....)).......((.((((..(((((((((..((...((((((..(...-----...)..)))))).))...)))))))))..)))).))))))).......... ( -37.60) >consensus CUGCUAAUCAUGGCAUUUGCUCGUUUACAUUAUGUGGUCGGUUUUCAGUUUUUGCAGCUUGCGC_UGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUA .......(((((.((..(((.((.((((.....)))).))......(((...(((((((...(((((((((....)))))))))...)))))))))))))...))))))).......... (-25.26 = -27.54 + 2.28)

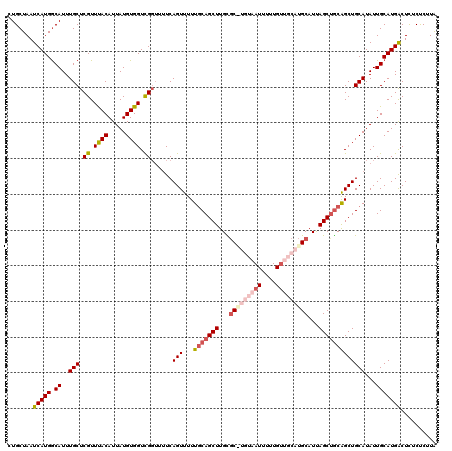
| Location | 21,098,646 – 21,098,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.96 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.84 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21098646 120 - 23771897 UAAGAGAGAGCCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAUCAAAAAUUACAUGCGCAAGCUGCAAAAACUGAAAACCGACCACAUAAUGUAAACGAGCAAAUGUCAUGAUUAGCAG .....(..(..((((((...(((...(((((((..((((((............))))))..)))))))............((...((.....))...)).)))..)).))))..)..).. ( -25.00) >DroSec_CAF1 148832 119 - 1 UAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACAUGCGCAAGCUG-AAAAACUGAAAACCGACCACAUAAUGUAAACGAGCAAAUGCCAUGAUUAGCAG .....(..(((((((((...(((.((((((.....)))).))........((((((((....))(.-...)...................))))))....)))..)).)))))))..).. ( -25.40) >DroSim_CAF1 56361 120 - 1 UAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACACGCGCAAGCUGCAAAAACUGAAAACCGACCACAUAAUGUAAACGAGCAAAUGCCAUGAUUAGCAG .....(..(((((((((...(((((((((.((....))..((..............)))).))))))).......................(((......)))..)).)))))))..).. ( -26.24) >DroEre_CAF1 71196 115 - 1 CAAGGGAAAGUCAUGCAAUAUGCAGCCGUGGCUGAUGCAUGCAACAAAAAU-----GCACAAGCUGCAAAAACUGAAAACCGACCACAUAAUGUAAACGAGCAAAUGCCAUGAUUAGCAG .....(..(((((((((...(((....(..(((..(((((.........))-----)))..)))..).............((...((.....))...)).)))..)).)))))))..).. ( -25.10) >DroYak_CAF1 43072 115 - 1 UAAGGGAAAGUCAUGCAAUAUGCAGCUGCAGCUGAUGCAUGCAACAAAAAU-----GAACAAGCUGCAAAAACUGAGCUGCAACCACAUAAUGUGAACGAGCAAAUGCCAUGAUUAGCAA .....(..(((((((((...(((((((((((((....(((.........))-----)....))))))........)))))))..(((.....)))..........)).)))))))..).. ( -31.40) >consensus UAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACA_GCGCAAGCUGCAAAAACUGAAAACCGACCACAUAAUGUAAACGAGCAAAUGCCAUGAUUAGCAG .....(..(((((((((...((((((((((((....)).)))...................))))))).......................(((......)))..)).)))))))..).. (-19.56 = -19.84 + 0.28)


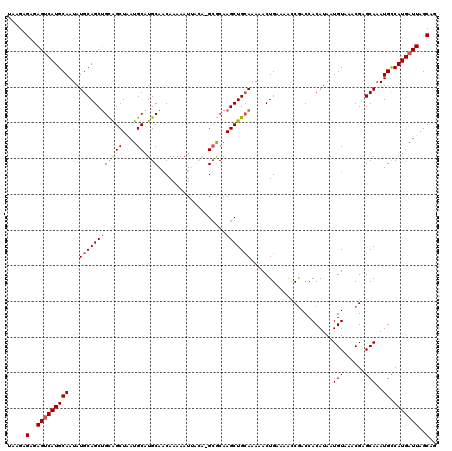
| Location | 21,098,686 – 21,098,806 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -29.99 |
| Consensus MFE | -22.80 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21098686 120 + 23771897 GUUUUCAGUUUUUGCAGCUUGCGCAUGUAAUUUUUGAUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGGCUCUCUCUUAGAAUUGCUUUCACCGCUGCACUUUUGGGAUUUAACAUUUU ............(((((((...(((((((........)))))))...)))))))(((((.....))..)))....((((((((.(((..........))).))))))))........... ( -32.10) >DroSec_CAF1 148872 119 + 1 GUUUUCAGUUUUU-CAGCUUGCGCAUGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUAGAAAUGCUUUCAUCGCUGCACUUUUGGGAUUUAACAUUUU ..(..(((.....-.((((...(((((((((....)))))))))...))))(((((........((((...(((.....))).)))).......)))))....)))..)........... ( -30.46) >DroSim_CAF1 56401 120 + 1 GUUUUCAGUUUUUGCAGCUUGCGCGUGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUAGAAAUGCUUUCACCGCUGCACUUUUGGGAUUUAACAUUUU .......((..((((((((...(((((((((....)))))))))...))))))))..))................((((((((.(((..........))).))))))))........... ( -34.20) >DroEre_CAF1 71236 115 + 1 GUUUUCAGUUUUUGCAGCUUGUGC-----AUUUUUGUUGCAUGCAUCAGCCACGGCUGCAUAUUGCAUGACUUUCCCUUGGAAAUGCUUUCACCACUGCACUUUUGGGAUUUUACAUUUU ..(..(((....(((((..((.((-----((((..(..(((((((.((((....)))).....)))))).).......)..))))))...))...)))))...)))..)........... ( -27.70) >DroYak_CAF1 43112 115 + 1 CAGCUCAGUUUUUGCAGCUUGUUC-----AUUUUUGUUGCAUGCAUCAGCUGCAGCUGCAUAUUGCAUGACUUUCCCUUAGAAAUGUUUUCACCACUGCACUUUUGGGAUUUUACAUUUU ...(((((....(((((..((..(-----((((((((((((.((....))))))))(((.....)))............)))))))....))...)))))...)))))............ ( -25.50) >consensus GUUUUCAGUUUUUGCAGCUUGCGC_UGUAAUUUUUGUUGCAUGCAUUAGCUGCAGCUGCAUAUUGCAUGACUCUCUCUUAGAAAUGCUUUCACCGCUGCACUUUUGGGAUUUAACAUUUU ......((((..(((((..((((((((((((....)))))))))...(((....))))))..))))).))))....(((((((.(((..........))).)))))))............ (-22.80 = -24.60 + 1.80)


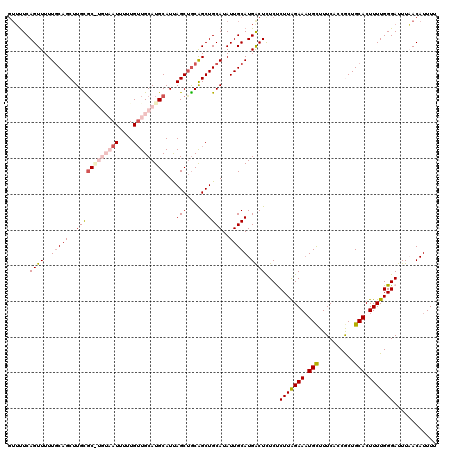
| Location | 21,098,686 – 21,098,806 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -21.86 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21098686 120 - 23771897 AAAAUGUUAAAUCCCAAAAGUGCAGCGGUGAAAGCAAUUCUAAGAGAGAGCCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAUCAAAAAUUACAUGCGCAAGCUGCAAAAACUGAAAAC ....................((((((.((....))..............((((((.((((((((..((((.....))))))))).....))).)))).))..))))))............ ( -26.30) >DroSec_CAF1 148872 119 - 1 AAAAUGUUAAAUCCCAAAAGUGCAGCGAUGAAAGCAUUUCUAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACAUGCGCAAGCUG-AAAAACUGAAAAC ..................((((((((.((((.....((((.....)))).))))((.....)).)))))((((..((((((.((......)).))))))..)))).-....)))...... ( -25.60) >DroSim_CAF1 56401 120 - 1 AAAAUGUUAAAUCCCAAAAGUGCAGCGGUGAAAGCAUUUCUAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACACGCGCAAGCUGCAAAAACUGAAAAC ....................((((((..((...((.((((.....))))(.((((((...(((....))).....)))))))..............)).)).))))))............ ( -26.20) >DroEre_CAF1 71236 115 - 1 AAAAUGUAAAAUCCCAAAAGUGCAGUGGUGAAAGCAUUUCCAAGGGAAAGUCAUGCAAUAUGCAGCCGUGGCUGAUGCAUGCAACAAAAAU-----GCACAAGCUGCAAAAACUGAAAAC ....................((((((..((...((((((((...)))))(.((((((.....((((....)))).)))))))........)-----)).)).))))))............ ( -27.50) >DroYak_CAF1 43112 115 - 1 AAAAUGUAAAAUCCCAAAAGUGCAGUGGUGAAAACAUUUCUAAGGGAAAGUCAUGCAAUAUGCAGCUGCAGCUGAUGCAUGCAACAAAAAU-----GAACAAGCUGCAAAAACUGAGCUG ..................(((.((((.((((.....((((.....)))).))))......((((((((((((....)).)))..((....)-----)....)))))))...)))).))). ( -25.40) >consensus AAAAUGUUAAAUCCCAAAAGUGCAGCGGUGAAAGCAUUUCUAAGAGAGAGUCAUGCAAUAUGCAGCUGCAGCUAAUGCAUGCAACAAAAAUUACA_GCGCAAGCUGCAAAAACUGAAAAC ....................((((((..((...((.(((((...)))))(.((((((...(((....))).....)))))))..............)).)).))))))............ (-21.86 = -21.26 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:28 2006