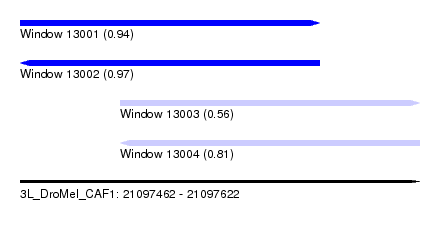
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,097,462 – 21,097,622 |
| Length | 160 |
| Max. P | 0.970956 |

| Location | 21,097,462 – 21,097,582 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21097462 120 + 23771897 CUGCAAUCCGAGUGGUCCUUGCCUUUGUUUAUCCUGCUGCCAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCCUACUGUCCUGCAGUUCCAGUUACU (((((....(((.(((....)))...((.(((((((....))))))).)))))(((......))).....((((...(((((((....)))))))....)))).)))))........... ( -32.90) >DroSec_CAF1 147654 120 + 1 CUGCAAUCCGAGUGGUCCUUGCCUUUGUUUAUCCUGCUGCCAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACU (((((....(((.(((....)))...((.(((((((....))))))).)))))(((......))).....((((...(((((((....)))))))....)))).)))))........... ( -33.60) >DroSim_CAF1 55186 120 + 1 CUGCAAUCCGAGUGGUCCUUGCCUUUGUUUAUCCUGCUGCCAGGAUGCACCUCGCACAUCCUUGUACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACU (((((...((((.(((...(((....((.(((((((....))))))).))...))).)))))))......((((...(((((((....)))))))....)))).)))))........... ( -33.50) >DroEre_CAF1 70017 120 + 1 CUGCAAUGCGAGUGGUCCUUGCCUUUGUUUAUCCUGCUGCCAUGUUGCAGCUCGCUUAUCCUUGCACCCUGGUACUUGCGCUUAUGAAUAAGCGUUUACUGUCCUGCAGUUCCAGUUACU (((((.((((((.(((....)))............(((((......))))).........))))))....((((...(((((((....))))))).))))....)))))........... ( -31.90) >DroYak_CAF1 41876 120 + 1 CUGCAAUCCGAGUGCUCCUUGCCUUUGUUUAUCCUGCUGCCAGGUUGCACCUUGCAUAUCCUUGCACUCUGACACUCGCGCUUAUGAAUAAGCGUUUACUGUCCUGCAGUUCCAGUUACU (((((....((((((....(((...(((....((((....))))..)))....))).......)))))).((((...(((((((....)))))))....)))).)))))........... ( -34.10) >consensus CUGCAAUCCGAGUGGUCCUUGCCUUUGUUUAUCCUGCUGCCAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACU (((((...((((.(((...(((....((.(((((((....))))))).))...))).)))))))......((((...(((((((....)))))))....)))).)))))........... (-26.80 = -27.44 + 0.64)

| Location | 21,097,462 – 21,097,582 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -31.06 |
| Energy contribution | -32.90 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21097462 120 - 23771897 AGUAACUGGAACUGCAGGACAGUAGGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUGGCAGCAGGAUAAACAAAGGCAAGGACCACUCGGAUUGCAG .((((((((..((((..((((.(..(((((((....))))))).).))))....(((.((((((..(...)..)))))).)))))))..........((......))..)))).)))).. ( -42.80) >DroSec_CAF1 147654 120 - 1 AGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUGGCAGCAGGAUAAACAAAGGCAAGGACCACUCGGAUUGCAG ...........((((((..((((..(((((((....)))))))...((((..(.(((.((((((..(...)..)))))).))).)..))))......((......))))).)..)))))) ( -42.60) >DroSim_CAF1 55186 120 - 1 AGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUACAAGGAUGUGCGAGGUGCAUCCUGGCAGCAGGAUAAACAAAGGCAAGGACCACUCGGAUUGCAG .((((((((..((((..((((.(..(((((((....))))))).).))))...((...((((((..(...)..)))))).)).))))..........((......))..)))).)))).. ( -39.40) >DroEre_CAF1 70017 120 - 1 AGUAACUGGAACUGCAGGACAGUAAACGCUUAUUCAUAAGCGCAAGUACCAGGGUGCAAGGAUAAGCGAGCUGCAACAUGGCAGCAGGAUAAACAAAGGCAAGGACCACUCGCAUUGCAG ...........((((((....((...(((((((((....((....))((....))....))))))))).(((((......))))).......))....((.((.....)).)).)))))) ( -34.00) >DroYak_CAF1 41876 120 - 1 AGUAACUGGAACUGCAGGACAGUAAACGCUUAUUCAUAAGCGCGAGUGUCAGAGUGCAAGGAUAUGCAAGGUGCAACCUGGCAGCAGGAUAAACAAAGGCAAGGAGCACUCGGAUUGCAG ...........((((((((((.(...((((((....))))))..).)))).((((((.......(((.....))).(((.((....(......)....)).))).))))))...)))))) ( -37.30) >consensus AGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUGGCAGCAGGAUAAACAAAGGCAAGGACCACUCGGAUUGCAG .((((((((..((((..........(((((((....)))))))...((((((((((((.(......)....))))))))))))))))..........((......))..)))).)))).. (-31.06 = -32.90 + 1.84)


| Location | 21,097,502 – 21,097,622 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.66 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21097502 120 + 23771897 CAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCCUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGGCAAUUAUGCGCUUCUUAAAAAUAAA .((((((.........)))))).(((....((((...(((((((....)))))))....))))...((((.......))))((((.......)))).....)))................ ( -30.50) >DroSec_CAF1 147694 120 + 1 CAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGGCAAUUAUGCGCUUCUUAAAAAUAAA .((((((.........)))))).(((....((((...(((((((....)))))))....))))...((((.......))))((((.......)))).....)))................ ( -31.20) >DroSim_CAF1 55226 120 + 1 CAGGAUGCACCUCGCACAUCCUUGUACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGGCAAUUAUGCGCUUCUUAAAAAUAAA .((((((.........))))))........((((...(((((((....)))))))....))))..(((....(((...)))((((.......)))).....)))................ ( -29.20) >DroEre_CAF1 70057 120 + 1 CAUGUUGCAGCUCGCUUAUCCUUGCACCCUGGUACUUGCGCUUAUGAAUAAGCGUUUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGCCAAUUAUGCGCUUCUUAAAAAUAAA ((((((((((..(((........)).....((((...(((((((....))))))).)))))..))))))..........(((((........)))))..))))................. ( -27.30) >DroYak_CAF1 41916 120 + 1 CAGGUUGCACCUUGCAUAUCCUUGCACUCUGACACUCGCGCUUAUGAAUAAGCGUUUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGGCAAUUAUGCGCUUCCUAAAAAUAAA .(((..((....((((......))))....((((...(((((((....)))))))....))))..(((....(((...)))((((.......)))).....)))))..)))......... ( -30.50) >consensus CAGGAUGCACCUCGCACAUCCUUGCACCCUGACACUUGCGCUUAUGAAUAAGCGCUUACUGUCCUGCAGUUCCAGUUACUGGCCCAGUUACCGGGCAAUUAUGCGCUUCUUAAAAAUAAA .((((.((.....(((......))).....((((...(((((((....)))))))....))))..(((....(((...)))((((.......)))).....))))).))))......... (-25.50 = -25.66 + 0.16)


| Location | 21,097,502 – 21,097,622 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -30.32 |
| Energy contribution | -32.36 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21097502 120 - 23771897 UUUAUUUUUAAGAAGCGCAUAAUUGCCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAGGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUG ..............((((....(((((((((.((((...)))).)))))....))))((((.(..(((((((....))))))).).))))...)))).((((((..(...)..)))))). ( -42.70) >DroSec_CAF1 147694 120 - 1 UUUAUUUUUAAGAAGCGCAUAAUUGCCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUG ..............((((....(((((((((.((((...)))).)))))....))))((((.(..(((((((....))))))).).))))...)))).((((((..(...)..)))))). ( -41.90) >DroSim_CAF1 55226 120 - 1 UUUAUUUUUAAGAAGCGCAUAAUUGCCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUACAAGGAUGUGCGAGGUGCAUCCUG ..............(((((....)))(((((.((((...)))).)))))....))..((((.(..(((((((....))))))).).))))........((((((..(...)..)))))). ( -38.40) >DroEre_CAF1 70057 120 - 1 UUUAUUUUUAAGAAGCGCAUAAUUGGCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAAACGCUUAUUCAUAAGCGCAAGUACCAGGGUGCAAGGAUAAGCGAGCUGCAACAUG ..............((((...(((((((........))))))).((((...(....)..))))...(((((((((....((....))((....))....))))))))).)).))...... ( -33.10) >DroYak_CAF1 41916 120 - 1 UUUAUUUUUAGGAAGCGCAUAAUUGCCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAAACGCUUAUUCAUAAGCGCGAGUGUCAGAGUGCAAGGAUAUGCAAGGUGCAACCUG ........((((..((((....(((((((((.((((...)))).)))))...((((.((((.(...((((((....))))))..).))))....)))).......)))).))))..)))) ( -38.20) >consensus UUUAUUUUUAAGAAGCGCAUAAUUGCCCGGUAACUGGGCCAGUAACUGGAACUGCAGGACAGUAAGCGCUUAUUCAUAAGCGCAAGUGUCAGGGUGCAAGGAUGUGCGAGGUGCAUCCUG ..............((((......((((((...))))))......((((.((((.....))))..(((((((....))))))).....)))).)))).(((((((((...))))))))). (-30.32 = -32.36 + 2.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:24 2006