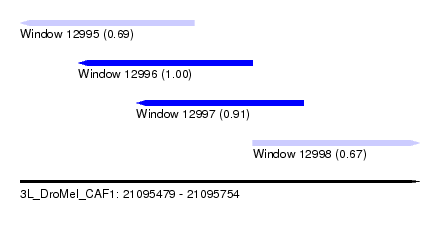
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,095,479 – 21,095,754 |
| Length | 275 |
| Max. P | 0.999921 |

| Location | 21,095,479 – 21,095,599 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21095479 120 - 23771897 AAGGCAAAACAUUUGACAUAUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACCGUUUAAAUGGCAAAGUAGGAAUGAAAAACACACACAAACC ........................(((((((((((...(((.((((..((......))...))).).))).)))))(.((((....)))))..............))))))......... ( -23.00) >DroSec_CAF1 145683 120 - 1 AAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAACGGCGACCGUUUAAAUGGCAAAGUAGGAGUGAAAAACACACACAAACC ((((((((.((((.((....)).)))))))))))).((((...(....((......)).(((((.(.((....)).).((((....))))........))))).....)...)))).... ( -27.80) >DroSim_CAF1 53187 120 - 1 AAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACCGUUUAAAUGGCAAAGUAGGAGUGAAAAACACACACAAACC ((((((((.((((.((....)).)))))))))))).((((...(....((......)).(((((.(.((....)).).((((....))))........))))).....)...)))).... ( -27.80) >DroEre_CAF1 68111 120 - 1 ACGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAAGAGCACCGAAAGCGGACACUCACACCAAAAGGCGACAGUUUAAAUGGCAAAGUAGGAGUGAUAAACACGUGCAAACC ...(((..((.((((.(((((..(.((((.(((((...(((..(((..(((....)))...)))...))).))))))))).)..))))).))))))....(((.....))).)))..... ( -29.70) >DroYak_CAF1 39918 120 - 1 AAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACAGUUUAAAUGGCAAAGCAGGAGUGAAAAACACACACAAGCC ..(((......((((.(((((..(.((((.(((((...(((.((((..((......))...))).).))).))))))))).)..))))).))))......(((.....)))......))) ( -29.80) >consensus AAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACCGUUUAAAUGGCAAAGUAGGAGUGAAAAACACACACAAACC ((((((((.((((.((....)).)))))))))))).((((........((......)).(((((.(.((....)).)...(((.....))).......))))).....))))........ (-24.22 = -24.46 + 0.24)

| Location | 21,095,519 – 21,095,639 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -36.88 |
| Energy contribution | -37.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21095519 120 - 23771897 UACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUAUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACC ....((((....(((((((((...(((((.....)))))(((((((((.((((.((....)).)))))))))))))...)))))))))((......))................)))).. ( -39.50) >DroSec_CAF1 145723 120 - 1 UACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAACGGCGACC ....((((....(((((((((...(((((.....)))))(((((((((.((((.((....)).)))))))))))))...)))))))))((......))................)))).. ( -39.60) >DroSim_CAF1 53227 120 - 1 UACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACC ....((((....(((((((((...(((((.....)))))(((((((((.((((.((....)).)))))))))))))...)))))))))((......))................)))).. ( -39.60) >DroEre_CAF1 68151 120 - 1 CACUCCGCAUCCUGCUCCAUCUGGGCAUGGCACACAUGUUACGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAAGAGCACCGAAAGCGGACACUCACACCAAAAGGCGACA .....(((....(((.((....)))))(((..((((((....((((((.((((.((....)).)))))))))).))))))...(((..(((....)))...)))...)))....)))... ( -32.80) >DroYak_CAF1 39958 120 - 1 UACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACA ....((((....(((((((((...(((((.....)))))(((((((((.((((.((....)).)))))))))))))...)))))))))((......))................)))).. ( -39.60) >consensus UACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUGGAGGAGCACCAAAAGCGGACACUCACACCAAAAGGCGACC ...(((((....(((((((((...(((((.....)))))(((((((((.((((.((....)).)))))))))))))...)))))))))......)))))......(.((....)).)... (-36.88 = -37.32 + 0.44)

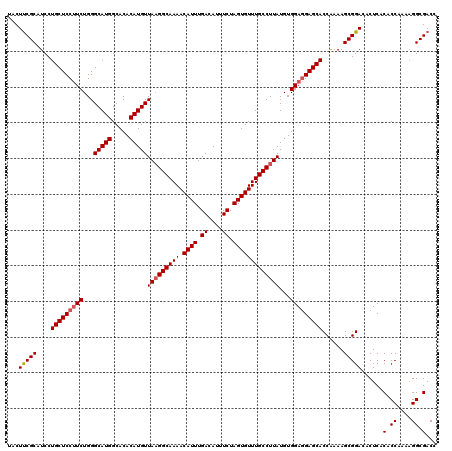
| Location | 21,095,559 – 21,095,674 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.34 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21095559 115 - 23771897 CCCGUGC----CGUCCUGCCGCUUUCCGCCUGAUCGUCCUACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUAUCUAGUGUUUGCCUUAUGUG ...((((----((...((((.......((..(((((........)).)))..))..(....))))))))))).(((((.((((((((.((((.((....)).))))))))))))))))) ( -32.50) >DroSec_CAF1 145763 119 - 1 CCCGUCCCGCCCGUCUUGCCCCUCUCCGCCUGAUCGUCCUACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUG ............(((.(((((......((..(((((........)).)))..)).......))))).))).((((...(((((((((.((((.((....)).))))))))))))))))) ( -32.12) >DroEre_CAF1 68191 107 - 1 -----------UGUCCUGCUCCUCG-CCCCGGAUCGUCCCACUCCGCAUCCUGCUCCAUCUGGGCAUGGCACACAUGUUACGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUG -----------.............(-((..((((((........)).))))(((.((....))))).))).((((((....((((((.((((.((....)).)))))))))).)))))) ( -29.00) >DroYak_CAF1 39998 108 - 1 -----------UGUCCUGCCGCUCUUCCCCUGAUCGUCCUACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUG -----------.....(((((..........(((((........)).))).(((.((....))))))))))((((...(((((((((.((((.((....)).))))))))))))))))) ( -28.10) >consensus ___________CGUCCUGCCCCUCUCCCCCUGAUCGUCCUACUUCGCAUCCUGCUCCUUCUGGGCAUGGCACACAUGUUAAGGCAAAACAUUUGACAUUUCUAGUGUUUGCCUUAUGUG ................((((...........(((((........)).))).(((.((....))))).))))((((((...(((((((.((((.((....)).))))))))))))))))) (-26.24 = -26.30 + 0.06)

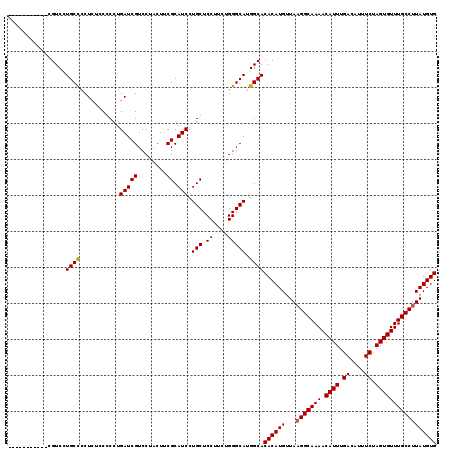
| Location | 21,095,639 – 21,095,754 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -30.14 |
| Energy contribution | -31.08 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21095639 115 + 23771897 GGACGAUCAGGCGGAAAGCGGCAGGACG----GCACGGGAACGGACAGGAGUAGCAGUCAGGGACAUUUGUCGUGCUGUUGGCUGUUUUUUAUGUCCUGCCCAUUUCCAUAGCAAUUGG ...((((...(((((((..(((((((((----...((....))(((((...(((((((..(.(((....))).)))))))).))))).....)))))))))..)))))...)).)))). ( -40.80) >DroSec_CAF1 145843 119 + 1 GGACGAUCAGGCGGAGAGGGGCAAGACGGGCGGGACGGGAAAGGACAGGAGUAGCAGUCAGGGACAUUUGUCGUGCUGUUGGCUGUUUUUUAUGUCCUGCCCAUUUCCAUAGCAAUUGG ...((((...(((((((..(......)((((((((((..(((((((((...(((((((..(.(((....))).)))))))).))))))))).)))))))))).)))))...)).)))). ( -44.70) >DroEre_CAF1 68271 104 + 1 GGACGAUCCGGGG-CGAGGAGCAGGACA--------------GGACGGGAGUAGCAGUCAGGGACAUUUGUCGUGCUGUUGGCUGUUUUUUAUGUCCUGCCCAUUUCCAUAGCAAUUGG ((.....))...(-(..((((..((.((--------------(((((.(((.(((((((((.(.(((.....))).).))))))))).))).))))))).))..))))...))...... ( -35.60) >DroYak_CAF1 40078 105 + 1 GGACGAUCAGGGGAAGAGCGGCAGGACA--------------UGUCGGGAGAAGCAGUCAGGGACAUUUGUCGUGCUGUUGGCUGUUUUUUAUGUCCUGCCCAUUUCCAUAGCAAUUGG ...((((....(((((.(.(((((((((--------------(.....(((((((((((((.(.(((.....))).).)))))))))))))))))))))))).)))))......)))). ( -40.50) >consensus GGACGAUCAGGCGGAGAGCGGCAGGACA______________GGACAGGAGUAGCAGUCAGGGACAUUUGUCGUGCUGUUGGCUGUUUUUUAUGUCCUGCCCAUUUCCAUAGCAAUUGG ...((((.....(((((.((((((((((................((....))(((((((((.(.(((.....))).).))))))))).....)))))))))).)))))......)))). (-30.14 = -31.08 + 0.94)


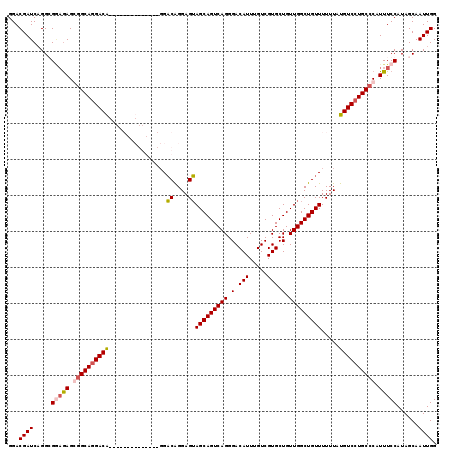
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:18 2006