
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,093,734 – 21,093,931 |
| Length | 197 |
| Max. P | 0.993362 |

| Location | 21,093,734 – 21,093,853 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21093734 119 - 23771897 UAGACUUGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGAGAAAAAUGCCAGAAAAUGUGGACAGAUCAAACAUUAUUUUACUACAUUCAAGUGCCUGGGAAAUUGCACA ..((.(((((.(((((...-...))))).))))).))....((((((((((((.((((...((...((....))..))...)))).))))..)))))))).((((..........)))). ( -22.60) >DroSec_CAF1 143974 119 - 1 UAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAAACAUUAUUUUACUACAUUCAAGUGCCUGGAAAAUUGCACA ..((.(((((.(((((...-...))))).))))).))....(((((((((((((((((...((...((....))..))...)))).)))))).))))))).((((..........)))). ( -28.10) >DroSim_CAF1 51024 119 - 1 UAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAAACAUUAUUUUACUACAUUCAAGUGCCUGGGAAAUUGCACA ..((.(((((.(((((...-...))))).))))).))....(((((((((((((((((...((...((....))..))...)))).)))))).))))))).((((..........)))). ( -28.10) >DroEre_CAF1 66360 120 - 1 UAGACUCGUUCGAGGGGAAAAAACCUUUAAGCGAUUCCAAAUGAGUGUGUGAACAAUGUCAGAAAAUGUGGGCAGAUCAAACAUUAUUUUACUACAUUCAAGUGUCUGUUAAAUUGCACA ((((((((((..((((.......))))..))))).......((((((((((((.(((((..((...((....))..))..)))))..))))).)))))))...)))))............ ( -30.10) >DroYak_CAF1 38177 119 - 1 UAGACUCGUUUAAGGGGAAAAAACCUUUAAACGACUCCAAAUGAAUGUGUGAAAUAUGUCAGAAAAUGUGAGCAGAUCAAACAUUAUUUUUCUACAUUCAAGUG-CUAGGAAAUUGCACA ..((.(((((((((((.......))))))))))).))....((((((((.((((.((((..((...((....))..))..))))....)))))))))))).(((-(.........)))). ( -34.30) >consensus UAGACUCGUUCAAGGGGAA_AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAAACAUUAUUUUACUACAUUCAAGUGCCUGGGAAAUUGCACA ..((.(((((.(((((.......))))).))))).))....(((((((((((((((((...((...((....))..))...)))).)))))).))))))).((((..........)))). (-24.36 = -24.60 + 0.24)



| Location | 21,093,774 – 21,093,893 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.16 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.65 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21093774 119 - 23771897 GACAAAGAUGCUGUUGAGAGAACUCAAGUUCUUAGCUCAAUAGACUUGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGAGAAAAAUGCCAGAAAAUGUGGACAGAUCAA ......(((.((((((((....)))...(((((((((((...((.(((((.(((((...-...))))).))))).))....))))).)))))).................)))))))).. ( -27.80) >DroSec_CAF1 144014 116 - 1 GACAAAGAUGCUGUCUU---CUCUUAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAA ......(((.((((((.---..(.....((((..(((((...((.(((((.(((((...-...))))).))))).))....)))))((((.....)))).))))...).))))))))).. ( -28.20) >DroSim_CAF1 51064 116 - 1 GACAAAGAUGCUGUCUU---CUCUCAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAA ......(((.((((((.---....((..((((..(((((...((.(((((.(((((...-...))))).))))).))....)))))((((.....)))).))))..)).))))))))).. ( -29.70) >DroEre_CAF1 66400 116 - 1 GACAAAUAUGCUGUCUU---CUC-CAAGUUCUUAGCUCAAUAGACUCGUUCGAGGGGAAAAAACCUUUAAGCGAUUCCAAAUGAGUGUGUGAACAAUGUCAGAAAAUGUGGGCAGAUCAA ..........((((((.---...-...((((...(((((...((.(((((..((((.......))))..))))).))....)))))....)))).....((.....)).))))))..... ( -28.20) >DroYak_CAF1 38216 117 - 1 GACAAAGAUGCUGUCUU---CACUCAAGUUCUAAGCUCAAUAGACUCGUUUAAGGGGAAAAAACCUUUAAACGACUCCAAAUGAAUGUGUGAAAUAUGUCAGAAAAUGUGAGCAGAUCAA ......(((.((((..(---(((.....((((..........((.(((((((((((.......))))))))))).)).....((((((.....)))).))))))...))))))))))).. ( -28.40) >consensus GACAAAGAUGCUGUCUU___CUCUCAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA_AAGCCUUUAAACGACUCCAAAUGAGUGUGUGAAAAAUGCCAGAAAAUGUGGACAGAUCAA ......(((.((((((............(((...(((((...((.(((((.(((((.......))))).))))).))....)))))....)))................))))))))).. (-22.69 = -22.65 + -0.04)


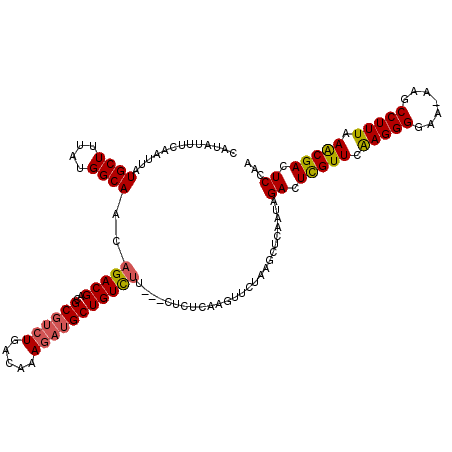
| Location | 21,093,814 – 21,093,931 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21093814 117 - 23771897 CAAAUUUCAAUUAUGCUUUAUGGCAACAGACGACGC--CUGACAAAGAUGCUGUUGAGAGAACUCAAGUUCUUAGCUCAAUAGACUUGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAA ..............(((((.((....)).....(.(--((((..(((...(((((((((((((....)))))...)))))))).)))..)).))).).)-))))................ ( -26.40) >DroSec_CAF1 144054 116 - 1 CAUAUUUCAAUUAUGCUUUAUGGCAGCAGACGACGCGUCUGACAAAGAUGCUGUCUU---CUCUUAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAA .........(((..((((.((...((.(((.(((((((((.....)))))).))).)---))))...))...))))..))).((.(((((.(((((...-...))))).))))).))... ( -28.70) >DroSim_CAF1 51104 116 - 1 CAUAUUUCAAUUAUGCUUUAUGGCAACAGACGACGCGUCUGACAAAGAUGCUGUCUU---CUCUCAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA-AAGCCUUUAAACGACUCCAA .........(((..((((..((....))((.(((((((((.....)))))).))).)---)...........))))..))).((.(((((.(((((...-...))))).))))).))... ( -28.10) >DroEre_CAF1 66440 116 - 1 CAUAUUUCAAUUAUGCUUUAUGGCAACAGACGUCGCGUCUGACAAAUAUGCUGUCUU---CUC-CAAGUUCUUAGCUCAAUAGACUCGUUCGAGGGGAAAAAACCUUUAAGCGAUUCCAA (((((((......((((....)))).((((((...))))))..)))))))..((((.---...-..(((.....)))....))))(((((..((((.......))))..)))))...... ( -28.00) >DroYak_CAF1 38256 117 - 1 CAUAUUUCAAUUAUGCUUUAUGGCAACAGACGUCGCGUCUGACAAAGAUGCUGUCUU---CACUCAAGUUCUAAGCUCAAUAGACUCGUUUAAGGGGAAAAAACCUUUAAACGACUCCAA .........(((..((((..((....))((((..((((((.....))))))))))..---............))))..))).((.(((((((((((.......))))))))))).))... ( -30.70) >consensus CAUAUUUCAAUUAUGCUUUAUGGCAACAGACGACGCGUCUGACAAAGAUGCUGUCUU___CUCUCAAGUUCUAAGCUCAAUAGACUCGUUCAAGGGGAA_AAGCCUUUAAACGACUCCAA .............((((....))))..(((((..((((((.....)))))))))))..........................((.(((((.(((((.......))))).))))).))... (-22.14 = -22.30 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:12 2006