
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,091,927 – 21,092,125 |
| Length | 198 |
| Max. P | 0.999940 |

| Location | 21,091,927 – 21,092,047 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21091927 120 + 23771897 AUAUUCGGCACAGGUCGGGAAAUGGCAAUCAUCAGAAUGACAGCUCGGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACUGCAAAGACAUCGUCCAUUUCUGUCAUGGAUAUGGCC ...((((((....))))))....(((....(((...(((((((...(((.(((((...(..........).)))))......((....))......)))....))))))).)))...))) ( -29.70) >DroSec_CAF1 142138 120 + 1 AUAUCCGGCACAGGUCAGGAAAUGGCAAUCAUCAGAAUGACAGCUCCGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCC (((((((.....((((.(((..((.((.((....)).)).))..)))...)))).........((((.(((((((((((((........)).)))))))))))))))..))))))).... ( -38.40) >DroSim_CAF1 49194 120 + 1 AUAUCCGGCACAGGUCAGGAAAUGGCAAUCAUCAGAAUGACAGCUCCUAAGGCCAAAACCAACGCAGCAGGUGGCUAAUUCACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCC ((((((((....((((((((..((.((.((....)).)).))..))))..))))....))...((((.(((((((((((((........)).)))))))))))))))...)))))).... ( -37.30) >DroEre_CAF1 64394 120 + 1 AUAUCCGGCACAGGUCAGGGAACGGCAAUCAUCAGAAUGACAACUCAGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCC (((((((((....(((.(....))))..((((....))))...........))).........((((.((((((((.((((........)).)).))))))))))))...)))))).... ( -30.60) >DroYak_CAF1 36270 120 + 1 AUAUCCGGCACAGGUCGGGAAAUGGCAAUAAUCAGAAUGACAGCUCAGAAGGCCAAUACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCC ...((((((....))))))....(((....(((...(((((((...(((.((((............((.(((((.....)))))))...(....))))).)))))))))).)))...))) ( -36.80) >consensus AUAUCCGGCACAGGUCAGGAAAUGGCAAUCAUCAGAAUGACAGCUCAGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCC (((((((((....))).(((((((((.....((.........(((.....))).............((.(((((.....)))))))...)).....))))))))).....)))))).... (-27.40 = -27.52 + 0.12)


| Location | 21,091,967 – 21,092,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -42.84 |
| Consensus MFE | -38.94 |
| Energy contribution | -39.78 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
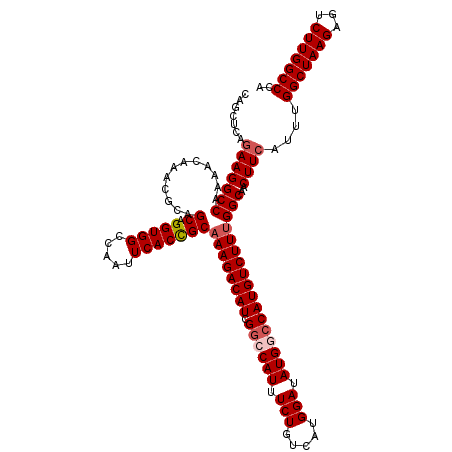
>3L_DroMel_CAF1 21091967 120 + 23771897 CAGCUCGGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACUGCAAAGACAUCGUCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCA ..........((((((.((...(((.....)))(((((.....(((((((((((.(.((((.(((.....))).))))).)))))))).))).......))))).....)).)))))).. ( -38.40) >DroSec_CAF1 142178 120 + 1 CAGCUCCGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCA .......((((((((((......((((.(((((((((((((........)).)))))))))))))))((((((....))))))..))))))..))))....(((((((...))))))).. ( -44.90) >DroSim_CAF1 49234 120 + 1 CAGCUCCUAAGGCCAAAACCAACGCAGCAGGUGGCUAAUUCACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCCAUGUCUUUGGCUACUUCAUUUGGCUAAGAGUCUUGGCCCA ..........(((((((......((((.(((((((((((((........)).)))))))))))))))((((((....))))))..))))))).........(((((((...))))))).. ( -40.60) >DroEre_CAF1 64434 120 + 1 CAACUCAGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUGGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCA .......(((((((............((.(((((.....))))))).(((((((.((((((.(((.....))).))))))))))))).)))..))))....(((((((...))))))).. ( -44.70) >DroYak_CAF1 36310 120 + 1 CAGCUCAGAAGGCCAAUACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCA .......(((((((............((.(((((.....)))))))((((((((.((((((.(((.....))).)))))))))))))))))..))))....(((((((...))))))).. ( -45.60) >consensus CAGCUCAGAAGGCCAAAACAAACGCAGCAGGUGGCCAAUUCACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCA .......(((((((............((.(((((.....)))))))((((((((.((((((.(((.....))).)))))))))))))))))..))))....(((((((...))))))).. (-38.94 = -39.78 + 0.84)


| Location | 21,091,967 – 21,092,087 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -39.04 |
| Energy contribution | -40.24 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21091967 120 - 23771897 UGGGCCAAGACUCUUAGCCAAAUGAAGUUGCCAAAGACAUGGCCAUAUCCAUGACAGAAAUGGACGAUGUCUUUGCAGUGAAUUGGCCACCUGCUGCGUUUGUUUUGGCCUUCCGAGCUG .((((((((((.(.((((..........(((.((((((((.(((((.((.......)).)))).).)))))))))))(((.......)))..)))).)...))))))))))......... ( -39.10) >DroSec_CAF1 142178 120 - 1 UGGGCCAAGACUCUUAGCCAAAUGAAGUUGCCAAAGACAUGGACAUAUCCAUGACAGAAAUGGCCAAUGUCUUUGCGGUGAAUUGGCCACCUGCUGCGUUUGUUUUGGCCUUCGGAGCUG .((((((((((.(.((((...........((.((((((((((.(((.((.......)).))).)).))))))))))((((.......)))).)))).)...))))))))))......... ( -39.30) >DroSim_CAF1 49234 120 - 1 UGGGCCAAGACUCUUAGCCAAAUGAAGUAGCCAAAGACAUGGACAUAUCCAUGACAGAAAUGGCCAAUGUCUUUGCGGUGAAUUAGCCACCUGCUGCGUUGGUUUUGGCCUUAGGAGCUG .(((((((((((.(((......))).((((((((((((((((.(((.((.......)).))).)).))))))))).((((.......)))).)))))...)))))))))))......... ( -41.20) >DroEre_CAF1 64434 120 - 1 UGGGCCAAGACUCUUAGCCAAAUGAAGUUGCCCAAGACAUGGCCAUAUCCAUGACAGAAAUGGCCGAUGUCUUUGCGGUGAAUUGGCCACCUGCUGCGUUUGUUUUGGCCUUCUGAGUUG .((((((((((.(.((((...........((..(((((((((((((.((.......)).)))))).))))))).))((((.......)))).)))).)...))))))))))......... ( -44.20) >DroYak_CAF1 36310 120 - 1 UGGGCCAAGACUCUUAGCCAAAUGAAGUUGCCAAAGACAUGGCCAUAUCCAUGACAGAAAUGGCCGAUGUCUUUGCGGUGAAUUGGCCACCUGCUGCGUUUGUAUUGGCCUUCUGAGCUG .(((((((.((.(.((((...........((.((((((((((((((.((.......)).)))))).))))))))))((((.......)))).)))).)...)).)))))))......... ( -42.80) >consensus UGGGCCAAGACUCUUAGCCAAAUGAAGUUGCCAAAGACAUGGCCAUAUCCAUGACAGAAAUGGCCGAUGUCUUUGCGGUGAAUUGGCCACCUGCUGCGUUUGUUUUGGCCUUCGGAGCUG .((((((((((.(.((((...........((.((((((((((((((.((.......)).)))))).))))))))))((((.......)))).)))).)...))))))))))......... (-39.04 = -40.24 + 1.20)



| Location | 21,092,007 – 21,092,125 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.72 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21092007 118 + 23771897 CACUGCAAAGACAUCGUCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAGGACGUUUCUUCCAGACAGAC--ACCCAUCCACCUA ..(((..((((...(((((.....(((((.((((((....)))))).))))).........(((((((...)))))))......)))))..)))).)))......--............. ( -31.90) >DroSec_CAF1 142218 118 + 1 CACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAGUCCGCUUCUUCCAGACAGAC--ACCCAUCCACCUA ....((.(((((.(((((((....(((((.((((((....)))))).)))))........)))))))..))))).)).......(((.(((......)).).)))--............. ( -29.20) >DroSim_CAF1 49274 118 + 1 CACCGCAAAGACAUUGGCCAUUUCUGUCAUGGAUAUGUCCAUGUCUUUGGCUACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAGUCCGCUUCUUCCAGACAGAC--ACCCAUCCACCUA ....((.(((((.(((((((.....((((.((((((....)))))).)))).........)))))))..))))).)).......(((.(((......)).).)))--............. ( -28.34) >DroEre_CAF1 64474 120 + 1 CACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUGGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAGGACGCUUCUUCUAGACAGCCAGAAGCAUUCACCUA ..((((.(((((((.((((((.(((.....))).)))))))))))))..))..........(((((((...)))))))......))..((((((.((....))..))))))......... ( -41.40) >DroYak_CAF1 36350 117 + 1 CACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAAGACGCUUCUUCCAG-CAGAC--AACCAUUCACCUA ....((((((((((.((((((.(((.....))).))))))))))))))(((..(((.....(((((((...))))))).....)))..))).......)-)....--............. ( -36.10) >consensus CACCGCAAAGACAUCGGCCAUUUCUGUCAUGGAUAUGGCCAUGUCUUUGGCAACUUCAUUUGGCUAAGAGUCUUGGCCCAUAAAGGACGCUUCUUCCAGACAGAC__ACCCAUCCACCUA .....(((((((((.((((((.(((.....))).)))))))))))))))((..(((.((..(((((((...))))))).)).)))...)).............................. (-27.88 = -28.72 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:04 2006