
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,081,103 – 21,081,383 |
| Length | 280 |
| Max. P | 0.815357 |

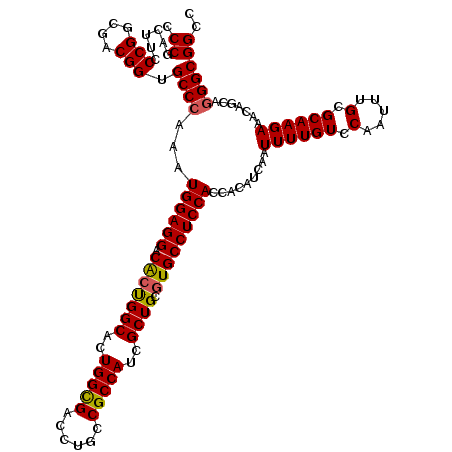
| Location | 21,081,103 – 21,081,223 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -42.76 |
| Energy contribution | -42.68 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21081103 120 - 23771897 CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACAAUGGCUCUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAGGGCGGCC .((......(((....))).((((...((((((.((..(((..(((((......)))))..)))...)))))))).........((((((.(.....).)))))).......)))))).. ( -42.00) >DroSec_CAF1 136453 120 - 1 CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGUGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAGGGCGGCC .((......(((....))).((((...((((((.(((((((..(((((......)))))..)))).))))))))).........((((((.(.....).)))))).......)))))).. ( -42.60) >DroSim_CAF1 42559 120 - 1 CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAUGGCGGCC ...........((((.(((((((....((.....))..))))))).)).)).((((((((.((((.(((.......))).....((((((.(.....).))))))..)))))))))))). ( -42.30) >DroEre_CAF1 58552 120 - 1 CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACGCCGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAGGGCGGCC .((......(((....))).((((...((((((.(((((((..(((((......)))))..)))).))))))))).........((((((.(.....).)))))).......)))))).. ( -47.00) >DroYak_CAF1 30634 120 - 1 CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAGGGCGGCC .((......(((....))).((((...((((((.(((((((..(((((......)))))..)))).))))))))).........((((((.(.....).)))))).......)))))).. ( -44.50) >consensus CCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACAUCAAUUUUGUCCAAUUUGCGCAAGAAACAGCAGGGCGGCC .((......(((....))).((((...((((((.(((((((..(((((......)))))..)))).))))))))).........((((((.(.....).)))))).......)))))).. (-42.76 = -42.68 + -0.08)


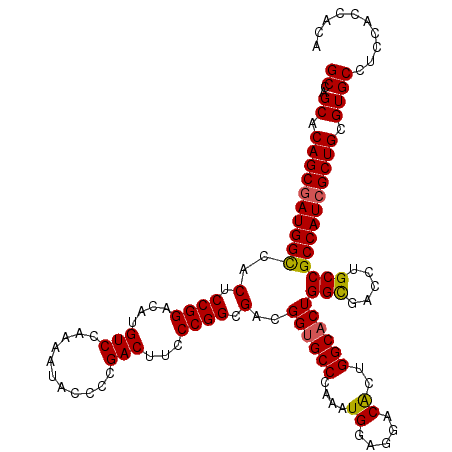
| Location | 21,081,143 – 21,081,263 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -40.16 |
| Energy contribution | -40.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21081143 120 - 23771897 GCCAGCACAGCAAUGGCCACUCCGGACAUGUCUAAAAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACAAUGGCUCUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ((.......))..(((.((((((((....(((...........)))...))))....)))).)))..((((((.((..(((..(((((......)))))..)))...))))))))..... ( -38.10) >DroSec_CAF1 136493 120 - 1 GCCAGCACAGCGAUGGUCACUCCGGACAUGUCCAAAAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGUGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ((..((.((((((((((..(((((((....)))....(((((((......)))....)))).......))))......((((..(....).)))))))))))))).)))).......... ( -43.10) >DroSim_CAF1 42599 120 - 1 GCCAGCACAGCGAUGGCCACUCCGGACAUGUCCAAAAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ((..((.((((((((((..(((((((....)))....(((((((......)))....)))).......))))......((((..(....).)))))))))))))).)))).......... ( -43.70) >DroEre_CAF1 58592 120 - 1 GCCGGCACAGCGAUGGCCACUCCGGACAUGUCCAACAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACGCCGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ...((((((((((((((..(.((((....(((...........)))...)))).)..(((..((....))(((.(((((....))))).)))))))))))))))..)))))......... ( -49.10) >DroYak_CAF1 30674 120 - 1 GCCAGCACAGCGAUGGCCACUCCGGACAUGUCGAAAAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ((..((.((((((((((..(.((((....((((.........))))...)))).)..((((((....((.....))..))))))(((.....))))))))))))).)))).......... ( -45.60) >consensus GCCAGCACAGCGAUGGCCACUCCGGACAUGUCCAAAAUACCCCGACUUCCCGGCGACGGUGCCCAAAUGGAGGACACUGGCACUGGCGACCUGCCGCCAUCGCUGCGUGCCUCCACCACA ((..((.((((((((((..(.((((....(((...........)))...)))).)..((((((....((.....))..))))))(((.....))))))))))))).)))).......... (-40.16 = -40.08 + -0.08)


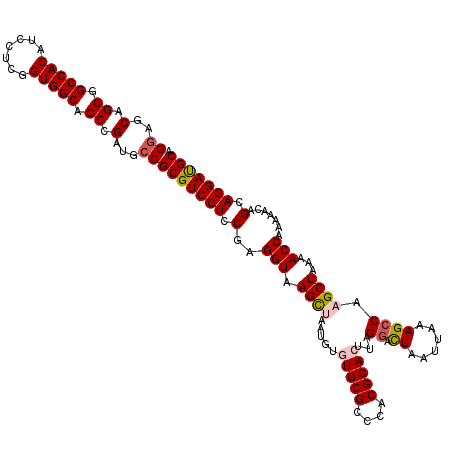
| Location | 21,081,263 – 21,081,383 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.76 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21081263 120 - 23771897 GGAGCAGCGGCCACAUCCUCAGUGGCAGCCGAUGCCGCGUCCUCCGAGGUAAGCUAAUGUGUGCGCCCACGCACUUAGGACUAAUUAAAGCCAAGCUAAAACCAAAAACAGCAGGAUGCA ((..(.((.(((((.......))))).)).)...))(((((((.(..(((.((((.....(((((....)))))...((.((......)))).))))...))).......).))))))). ( -38.90) >DroSec_CAF1 136613 120 - 1 GGAGCAGCGGCCACAUCCUCGGUGGCAGCCGAUGCCGCGUCCUCCGAGGUAAGCUAAUGUGUGCGCCCACGCACUUAGGAUUAAUUAAAGCCAAGCUAAAACCAAAAACAGCAGGAUGCA ((..(.((.(((((.......))))).)).)...))(((((((.(..(((.((((.....(((((....)))))...((...........)).))))...))).......).))))))). ( -36.70) >DroSim_CAF1 42719 120 - 1 GGAGCAGCGGCCACAUCCUCGGUGGCAGCCGAUGCCGCGUCCUCCGAGGUAAGCUAAUGUGUGCGCCCACGCACUUAGGACUAAUUAAAGCCAAGCUAAAACCAAAAACAGCAGGAUGCA ((..(.((.(((((.......))))).)).)...))(((((((.(..(((.((((.....(((((....)))))...((.((......)))).))))...))).......).))))))). ( -38.80) >DroEre_CAF1 58712 116 - 1 GGAGCAGCGGCCACAUCCUCGGUGGCAGCAGAUGCCGCGUCCUCCGAGGUAAGUCAAUG--UGCGCCCACGCACUUAGGACUAAUUAAAGACUAGCUAAAACCAA--ACAGCAGGACGCA ((..(.((.(((((.......))))).)).)...))(((((((....(((.((((...(--((((....)))))....)))).)))........(((........--..)))))))))). ( -39.70) >DroYak_CAF1 30794 118 - 1 GGAGCAGCGGCCACAUCCUCGGUGGCAGCAGAUGGCGCGUCCUCCGAGGUAAGUCAAUG--UGCGCCCACGCACUUAGGACUAAUUAAAGCCAAGCUAAAACCAAAAACAGCAGGAUGCA ...(((.((((.....((((((.(((.((.....))..).)).))))))..((((...(--((((....)))))....)))).......)))..(((............)))..).))). ( -37.40) >consensus GGAGCAGCGGCCACAUCCUCGGUGGCAGCCGAUGCCGCGUCCUCCGAGGUAAGCUAAUGUGUGCGCCCACGCACUUAGGACUAAUUAAAGCCAAGCUAAAACCAAAAACAGCAGGAUGCA ((..(.((.(((((.......))))).)).)...))(((((((.(..(((.((((.....(((((....)))))...((.((......)))).))))...))).......).))))))). (-32.12 = -32.76 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:55 2006