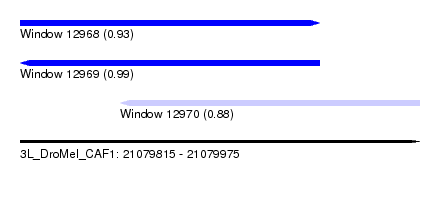
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,079,815 – 21,079,975 |
| Length | 160 |
| Max. P | 0.987009 |

| Location | 21,079,815 – 21,079,935 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -41.52 |
| Energy contribution | -41.68 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21079815 120 + 23771897 AAUGGCUGGCCAAUUACCUCGCCGUGCUGCAGUUUGAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACGCACCACUUCAGGGGCAGUGGCAGCAUCGCA ...(((..((...((((((.((((((.(((.(((....))).)))..)))))).))))))..))..)))((((((.(.((((((.................))))...)).).)))))). ( -46.53) >DroSec_CAF1 135165 120 + 1 GAUGGCAGGCCAAUUACCUCGCCGUGCUGCAGUUUGAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACGCACCACUUCAGGGGCAGUGGCAGCAUCGCA .((((((((.......))).)))))(((((...........((..(((((((((((.......)))))))))))....))((((.................))))....)))))...... ( -45.93) >DroSim_CAF1 41281 120 + 1 GAUGGCAGGCCAAUUACCUCGCCGUGCUGCAGUUUAAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACGCACCACUUCGGGGGCAGUGGGAGCAUCGCA ...((((((.......))).)))(((.(((...........((..(((((((((((.......)))))))))))....)).(((((...((.((.(....))))).))))).))).))). ( -50.10) >DroEre_CAF1 57373 120 + 1 GAUGGCAGGCCAAUUACCUCGCCGUGCUGCAGUUUGAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACACACCACUUUAGAGGCAGUGGCAGCAUCGCA .((((((((.......))).)))))(((((.(((((.((.(((..(((((((((((.......)))))))))))....)))..))))).))..((((........)))))))))...... ( -44.00) >DroYak_CAF1 29040 120 + 1 GGUGGAAGGCCAAUUACAUUGCCGUGCUGCAGUUUGAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACACACCACUUUAGAGGAAGUGGCAGCAUCGCA (((((((.((((((((.(((((......))))).))))..)))).(((((((((((.......)))))))))))..))))))).........(((((((....))))))).......... ( -45.80) >consensus GAUGGCAGGCCAAUUACCUCGCCGUGCUGCAGUUUGAUGAUGGUAAUUACGGCCAGGUGACAGCUGGCCGUGAUGGUUCCGCCCACAACGCACCACUUCAGGGGCAGUGGCAGCAUCGCA ..(((....)))........((.(((((((.(((((.((.(((..(((((((((((.......)))))))))))....)))..))))).))..((((........))))))))))).)). (-41.52 = -41.68 + 0.16)

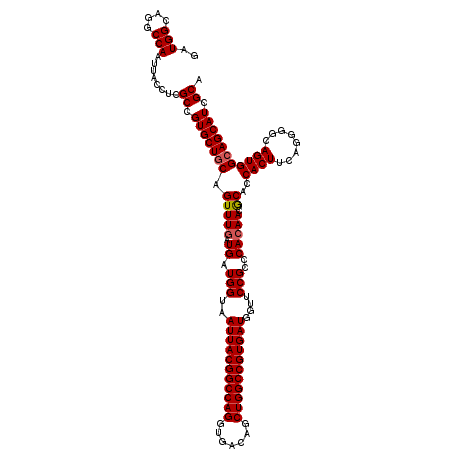
| Location | 21,079,815 – 21,079,935 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -41.30 |
| Energy contribution | -41.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21079815 120 - 23771897 UGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUCAAACUGCAGCACGGCGAGGUAAUUGGCCAGCCAUU .(((((((.((((((........))))))))))))).(((((..((...((((((((.......))))))))(((((((.((..(............)..))))))))))))).)))... ( -45.60) >DroSec_CAF1 135165 120 - 1 UGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUCAAACUGCAGCACGGCGAGGUAAUUGGCCUGCCAUC .(((((((.((((((........))))))))))))).(((((..((...((((((((.......))))))))(((((((.((..(............)..))))))))))).)))))... ( -47.70) >DroSim_CAF1 41281 120 - 1 UGCGAUGCUCCCACUGCCCCCGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUUAAACUGCAGCACGGCGAGGUAAUUGGCCUGCCAUC .(((((((..(((((........))))).))))))).(((((..((...((((((((.......))))))))(((((((.((........((......))))))))))))).)))))... ( -45.20) >DroEre_CAF1 57373 120 - 1 UGCGAUGCUGCCACUGCCUCUAAAGUGGUGUGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUCAAACUGCAGCACGGCGAGGUAAUUGGCCUGCCAUC .(((((((.((((((........))))))))))))).(((((..((...((((((((.......))))))))(((((((.((..(............)..))))))))))).)))))... ( -45.10) >DroYak_CAF1 29040 120 - 1 UGCGAUGCUGCCACUUCCUCUAAAGUGGUGUGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUCAAACUGCAGCACGGCAAUGUAAUUGGCCUUCCACC ((((.(((((((((((......)))))))((((((..(..(....)...((((((((.......))))))))................)..)))))))))).))))..(((....))).. ( -42.00) >consensus UGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCAUCAUCAAACUGCAGCACGGCGAGGUAAUUGGCCUGCCAUC .(((((((.((((((........))))))))))))).(((((..((...((((((((.......))))))))(((((((..........(((......))).))))))))).)))))... (-41.30 = -41.70 + 0.40)

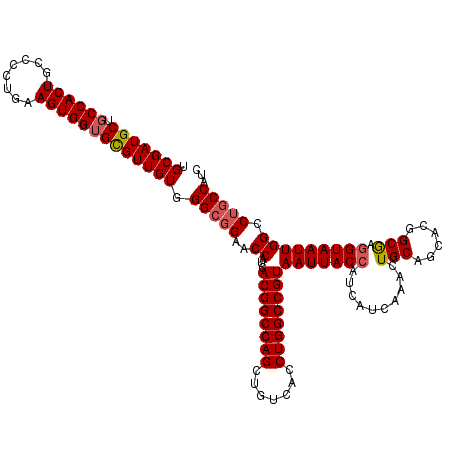
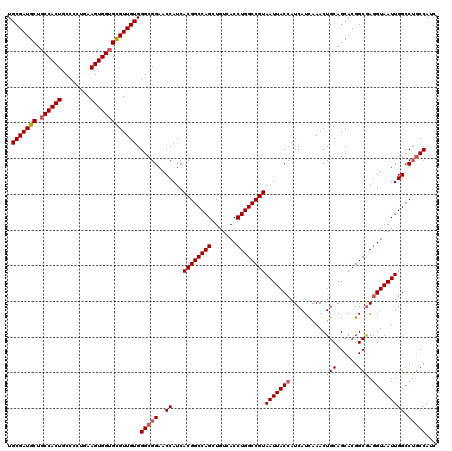
| Location | 21,079,855 – 21,079,975 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -41.26 |
| Energy contribution | -41.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21079855 120 - 23771897 ACCUGGUCUGGUACAUCCUGUUCGUCAUCUUCGUGCCAUAUGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA ....(((.((((...(((....((.......)).(((....(((((((.((((((........))))))))))))).))))))))))..((((((((.......))))))))....))). ( -43.90) >DroSec_CAF1 135205 120 - 1 ACCUGGUCUGGUACAUCCUGUUCGUCAUCUUCGUGCCGUAUGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA .........((((...................((.((((.((((((((.((((((........)))))))))))))).)))).))....((((((((.......))))))))...)))). ( -43.60) >DroSim_CAF1 41321 120 - 1 ACCUGGUCUGGUACAUCCUGUUCGUCAUCUUCGUGCCGUAUGCGAUGCUCCCACUGCCCCCGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA .........((((...................((.((((.((((((((..(((((........))))).)))))))).)))).))....((((((((.......))))))))...)))). ( -41.80) >DroEre_CAF1 57413 120 - 1 ACCUGGUCUGGUACAUCCUGUUCGUCAUCUUCGUGCCAUAUGCGAUGCUGCCACUGCCUCUAAAGUGGUGUGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA ....(((.((((...(((....((.......)).(((....(((((((.((((((........))))))))))))).))))))))))..((((((((.......))))))))....))). ( -41.30) >DroYak_CAF1 29080 120 - 1 ACCUGGUUUGGUACAUCCUGUUCGUCAUCUUCGUGCCGUAUGCGAUGCUGCCACUUCCUCUAAAGUGGUGUGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA ...(((((((((((....((.....)).....)))))((.((((((((.(((((((......))))))))))))))).)).))))))..((((((((.......))))))))........ ( -42.00) >consensus ACCUGGUCUGGUACAUCCUGUUCGUCAUCUUCGUGCCGUAUGCGAUGCUGCCACUGCCCCUGAAGUGGUGCGUUGUGGGCGGAACCAUCACGGCCAGCUGUCACCUGGCCGUAAUUACCA ....(((.((((...(((....((.......)).(((....(((((((.((((((........))))))))))))).))))))))))..((((((((.......))))))))....))). (-41.26 = -41.22 + -0.04)

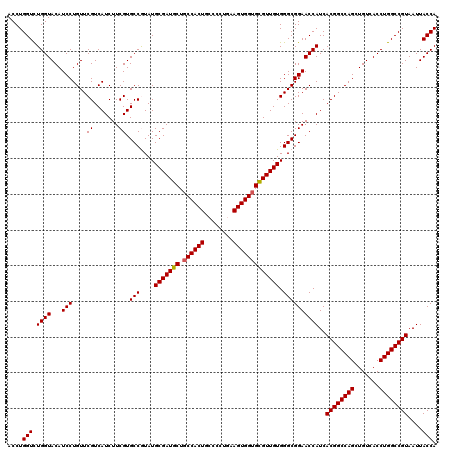
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:50 2006