
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,077,860 – 21,077,973 |
| Length | 113 |
| Max. P | 0.998541 |

| Location | 21,077,860 – 21,077,973 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -9.68 |
| Energy contribution | -12.64 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.42 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21077860 113 + 23771897 AUUU-CACCCUAUUGCCCUCAAAAGUAUGCUACACCUUUAAUGGAUUUUCCAUUUGCUCGCUAUACC-UUUUGUUUG-----UUUAAAUCAUUUAAAUAAAAUUUUGAGGGCCACACUUC ....-.........((((((((((((((((.........(((((.....))))).....)).)))).-.....((((-----(((((.....))))))))).))))))))))........ ( -25.84) >DroSec_CAF1 133135 113 + 1 AUUU-CAACCC-UUGCCCUCAAAAGUAUGCUACACCUUUAAUGGAUUUUCCAUUUGCUUGCUGUACCAUUUUGUUUG-----UUUAAAUCAUUUAAAUAACAUUUUGAGGGCCACACUUC ....-......-..((((((((((..(((.((((.(...(((((.....))))).....).)))).)))..((((.(-----(((((.....))))))))))))))))))))........ ( -26.60) >DroSim_CAF1 39422 113 + 1 AUUU-CAACCC-UUGCCCUCAAAAGUAUGCUACACCUUUAAUGGAUUUUCCAUUGGCUUGCUGUACCUUUUUGUUUG-----UUUAAAUCAUUUAAAUAACAUUUUGAGGGCCACACUUC ....-......-..((((((((((((((((....((...(((((.....)))))))...)).)))).....((((.(-----(((((.....))))))))))))))))))))........ ( -27.30) >DroEre_CAF1 55678 115 + 1 CAUUUAAACC--CCGCCCUCAAAAGUAUGCUACAGCUUAAAUGGUUUC-CC-GCUCUUUGCUGCACCACUUUGCUUGCUUAACUUAAAUCAUUUAAAUAACAU-GUUGGGGCCACACUUC ..........--..((((.(((..((..((....))(((((((((((.-..-((.....)).(((......)))...........)))))))))))...))..-.)))))))........ ( -20.80) >DroYak_CAF1 27118 109 + 1 AUUUCCAACC--CAGCCUUCC--AGUGUGCUACACCUUAAAUGCCCUCACC-UCUUUUCAUUGCACCAUUUUGCUU------UUUAAAUUAUUUAAAUAACUUUGUGAGGACCACACUUU ..........--.........--((((((......(......).((((((.-..........(((......)))..------(((((.....))))).......))))))..)))))).. ( -14.20) >consensus AUUU_CAACC__UUGCCCUCAAAAGUAUGCUACACCUUUAAUGGAUUUUCCAUUUGCUUGCUGUACCAUUUUGUUUG_____UUUAAAUCAUUUAAAUAACAUUUUGAGGGCCACACUUC ..............((((((((((((((((.......(((((((.....)))))))...)).))))................(((((.....))))).....))))))))))........ ( -9.68 = -12.64 + 2.96)

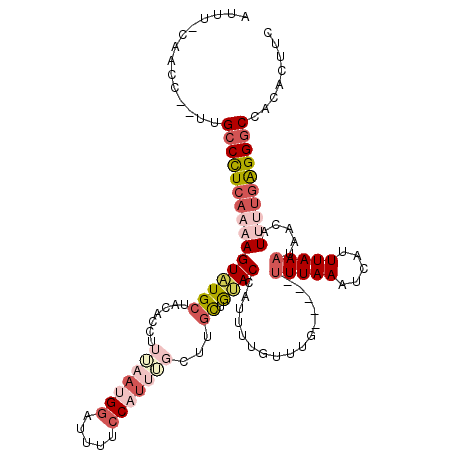
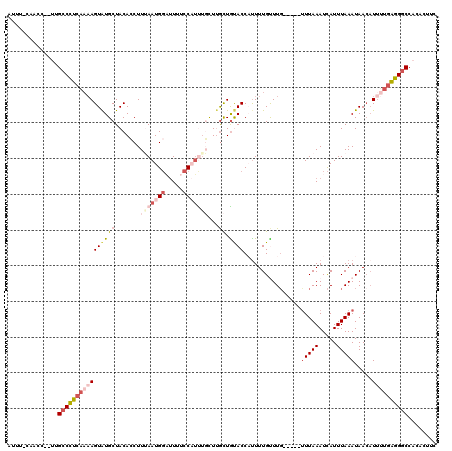
| Location | 21,077,860 – 21,077,973 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -14.90 |
| Energy contribution | -16.38 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21077860 113 - 23771897 GAAGUGUGGCCCUCAAAAUUUUAUUUAAAUGAUUUAAA-----CAAACAAAA-GGUAUAGCGAGCAAAUGGAAAAUCCAUUAAAGGUGUAGCAUACUUUUGAGGGCAAUAGGGUG-AAAU ....(((.((((((.(((((..........)))))...-----......(((-(((((.((..((.(((((.....)))))....))...)))))))))))))))).))).....-.... ( -26.40) >DroSec_CAF1 133135 113 - 1 GAAGUGUGGCCCUCAAAAUGUUAUUUAAAUGAUUUAAA-----CAAACAAAAUGGUACAGCAAGCAAAUGGAAAAUCCAUUAAAGGUGUAGCAUACUUUUGAGGGCAA-GGGUUG-AAAU ........((((((((((((((.(((((.....)))))-----..))))..(((.((((.(.....(((((.....)))))...).)))).)))..))))))))))..-......-.... ( -28.70) >DroSim_CAF1 39422 113 - 1 GAAGUGUGGCCCUCAAAAUGUUAUUUAAAUGAUUUAAA-----CAAACAAAAAGGUACAGCAAGCCAAUGGAAAAUCCAUUAAAGGUGUAGCAUACUUUUGAGGGCAA-GGGUUG-AAAU ........((((((....((((.(((((.....)))))-----..)))).(((((((..((..((((((((.....)))))...)))...)).)))))))))))))..-......-.... ( -27.30) >DroEre_CAF1 55678 115 - 1 GAAGUGUGGCCCCAAC-AUGUUAUUUAAAUGAUUUAAGUUAAGCAAGCAAAGUGGUGCAGCAAAGAGC-GG-GAAACCAUUUAAGCUGUAGCAUACUUUUGAGGGCGG--GGUUUAAAUG ...((..(((((((((-..(((((....)))))....)))..((...(((((((.((((((..(((..-((-....)).)))..)))))).)))...))))...))))--))))...)). ( -32.80) >DroYak_CAF1 27118 109 - 1 AAAGUGUGGUCCUCACAAAGUUAUUUAAAUAAUUUAAA------AAGCAAAAUGGUGCAAUGAAAAGA-GGUGAGGGCAUUUAAGGUGUAGCACACU--GGAAGGCUG--GGUUGGAAAU ..((((((..((((((.(((((((....)))))))...------..(((......)))..........-.))))))((((.....))))..))))))--.........--.......... ( -24.00) >consensus GAAGUGUGGCCCUCAAAAUGUUAUUUAAAUGAUUUAAA_____CAAACAAAAUGGUACAGCAAGCAAAUGGAAAAUCCAUUAAAGGUGUAGCAUACUUUUGAGGGCAA__GGUUG_AAAU ........((((((((((.(((((....)))))....................(.((((.(.....(((((.....)))))...).)))).)....)))))))))).............. (-14.90 = -16.38 + 1.48)

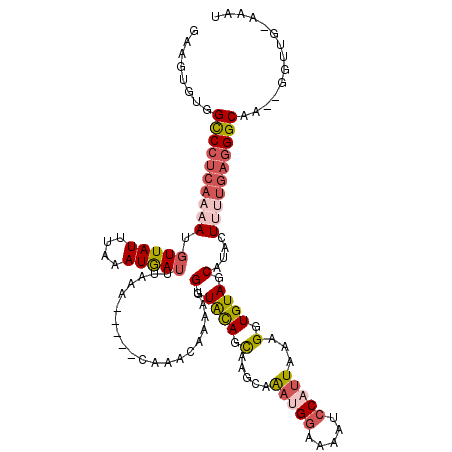
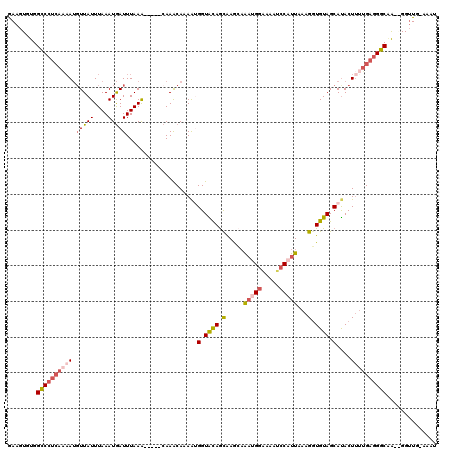
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:44 2006