
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,075,530 – 21,075,730 |
| Length | 200 |
| Max. P | 0.952198 |

| Location | 21,075,530 – 21,075,650 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -26.28 |
| Energy contribution | -27.28 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21075530 120 + 23771897 UUCUAUUAUUUGGUUUCUUGUGUCUAAACACGCCAAGCUUGCAAAUUCCCAAACAAUUGUUGUGAGCUACACAGGAGCAAAGAAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCG .(((.((((((((((((((((((....))))(((.((((..(((...............)))..))))......).)).)))))))))))))))))(((((...)))))........... ( -31.36) >DroSec_CAF1 130770 120 + 1 UUCUAUUAUUUGGUUUCUUGUGUCUUAACACGCCAAGCUUGCAAAUUCCCAAACAAUUGUUGUGAGCUACACAGGAGCAAAGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCG .(((.((((((((((((((((((....))))(((.((((..(((...............)))..))))......).)).)))))))))))))))))(((((...)))))........... ( -30.86) >DroSim_CAF1 37008 120 + 1 UUCUAUUAUUUGGUUUCUUGUGUCUAAACACGCCAAGCUUGCAAAUUCCCAAACAAUUGUUGUGAGCUACACAGGAGCAAAGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCG .(((.((((((((((((((((((....))))(((.((((..(((...............)))..))))......).)).)))))))))))))))))(((((...)))))........... ( -30.86) >DroEre_CAF1 53404 112 + 1 UUCUAUUAUUCGGUUUCUUGUGUCUAAACACGCCAGGCUUGCAAAUUCCCAAACAAUUGUUGCGAGCUACGCAGGA--------GCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCG .(((.(((((.(((((((.((((....))))((..(((((((((...............)))))))))..))))))--------))).))))))))(((((...)))))........... ( -29.46) >DroYak_CAF1 24894 120 + 1 UUCUAUUAUUUGGUUUCUUGUGUCUAAACACGCCAAGCUUGCAAAUUCCCGAACAAUUGUUGUGCGCUACACAGGAGCAAAGGAGCUAAAGAAAGAGCUUGGACUAAGCCACAGGACCCG ...........((..(((((((.((.......((((((((.(......((.(((....)))(((.....))).))(((......)))...)...))))))))....)).))))))).)). ( -28.40) >consensus UUCUAUUAUUUGGUUUCUUGUGUCUAAACACGCCAAGCUUGCAAAUUCCCAAACAAUUGUUGUGAGCUACACAGGAGCAAAGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCG .(((.((((((((((((((((((....))))....(((((((((...............)))))))))...........)))))))))))))))))(((((...)))))........... (-26.28 = -27.28 + 1.00)

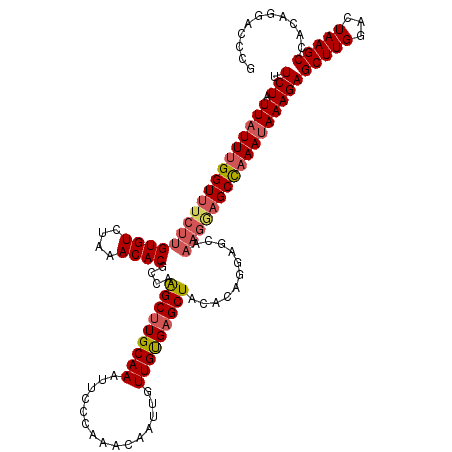

| Location | 21,075,610 – 21,075,730 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -17.98 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21075610 120 + 23771897 AGAAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCGUAAAUUGGCCAAUUAAAACUGCGGCCAAAAUUAAGAAGAAAAUUAAUUAAGACUUGCAGCCAAAGUCAAAGUUAUUUGAA ......((((((....((((.((((..((..((((.........((((((............))))))((((((........))))))....))))..))...)))).)))))))))).. ( -24.90) >DroSec_CAF1 130850 103 + 1 AGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCGCAAAUUGGCCAAUUAAAACUGCGGCCAGG-----------------CUGAGACUUGCAGCCAAAGUCGAAGUUAUUUGAA ......((((((....((((.((((..(((.(((.....((......)).........))).)))..((-----------------(((.......)))))..)))).)))))))))).. ( -28.34) >DroSim_CAF1 37088 103 + 1 AGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCGCAAAUUGGCCAAUUAAAACUGCGGCCGGG-----------------CUGAGACUUGCAACCAAAGUCAAAGUUAUUUGAA ......(((((((..(((((((......((...))..(((((((((....)))).....))))))))))-----------------))..(((((.......)))))....))))))).. ( -23.90) >DroEre_CAF1 53480 99 + 1 ----GCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCGCAAAUUGGCCAAUUAAAACUGUGGCCAGG-----------------CUGGGACUGGCAUCCAAAGUCAAAGUUAUUUGGA ----.((((((((.((..(((((....((((..(..((.((....((((((..........)))))).)-----------------).))..))))).)))))..))....)))))))). ( -33.60) >DroYak_CAF1 24974 102 + 1 AGGAGCUAAAGAAAGAGCUUGGACUAAGCCACAGGACCCGCAAAUUGGCCAAUUAAAACUGUGGCCAGG-----------------CUGAGACUA-CAGCCAAAGUCAAAGUUAUUUGAA ..(((((........))))).((((..(((((((.....((......)).........)))))))..((-----------------(((......-)))))..))))............. ( -29.14) >consensus AGGAGCCAAAUAAAGAGCUUGGACUAAGCCACAGGACCCGCAAAUUGGCCAAUUAAAACUGCGGCCAGG_________________CUGAGACUUGCAGCCAAAGUCAAAGUUAUUUGAA ......(((((((...((..(..((.......))..)..))...((((((............))))))......................(((((.......)))))....))))))).. (-17.98 = -17.94 + -0.04)

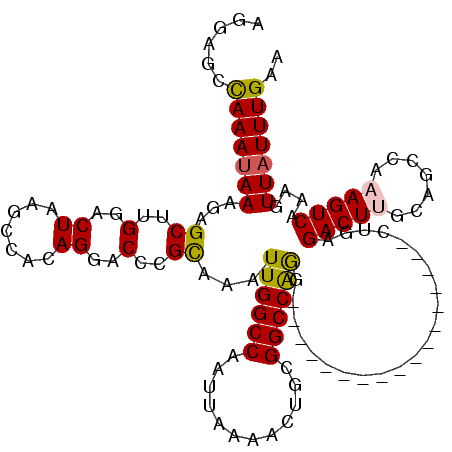

| Location | 21,075,610 – 21,075,730 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21075610 120 - 23771897 UUCAAAUAACUUUGACUUUGGCUGCAAGUCUUAAUUAAUUUUCUUCUUAAUUUUGGCCGCAGUUUUAAUUGGCCAAUUUACGGGUCCUGUGGCUUAGUCCAAGCUCUUUAUUUGGCUUCU .(((((((((((.((((..(((..((.(.(((((((((........))))))(((((((..........))))))).....))).).))..))).)))).)))....))))))))..... ( -28.80) >DroSec_CAF1 130850 103 - 1 UUCAAAUAACUUCGACUUUGGCUGCAAGUCUCAG-----------------CCUGGCCGCAGUUUUAAUUGGCCAAUUUGCGGGUCCUGUGGCUUAGUCCAAGCUCUUUAUUUGGCUCCU .(((((((((((.((((..(((..((.(.(((.(-----------------(.((((((..........))))))....))))).).))..))).)))).)))....))))))))..... ( -31.10) >DroSim_CAF1 37088 103 - 1 UUCAAAUAACUUUGACUUUGGUUGCAAGUCUCAG-----------------CCCGGCCGCAGUUUUAAUUGGCCAAUUUGCGGGUCCUGUGGCUUAGUCCAAGCUCUUUAUUUGGCUCCU .(((((((((((.((((..(((..((.(.(((.(-----------------(..(((((..........))))).....))))).).))..))).)))).)))....))))))))..... ( -27.90) >DroEre_CAF1 53480 99 - 1 UCCAAAUAACUUUGACUUUGGAUGCCAGUCCCAG-----------------CCUGGCCACAGUUUUAAUUGGCCAAUUUGCGGGUCCUGUGGCUUAGUCCAAGCUCUUUAUUUGGC---- .((((((((....((.((((((((((((.(((.(-----------------(.((((((..........))))))....))))).)...))))...))))))).)).)))))))).---- ( -33.70) >DroYak_CAF1 24974 102 - 1 UUCAAAUAACUUUGACUUUGGCUG-UAGUCUCAG-----------------CCUGGCCACAGUUUUAAUUGGCCAAUUUGCGGGUCCUGUGGCUUAGUCCAAGCUCUUUCUUUAGCUCCU .........(((.((((..(((((-......)))-----------------)).((((((((........((((........)))))))))))).)))).)))................. ( -28.40) >consensus UUCAAAUAACUUUGACUUUGGCUGCAAGUCUCAG_________________CCUGGCCGCAGUUUUAAUUGGCCAAUUUGCGGGUCCUGUGGCUUAGUCCAAGCUCUUUAUUUGGCUCCU .(((((((((((.((((..(((.....)))........................((((((((........((((........)))))))))))).)))).)))....))))))))..... (-22.42 = -22.50 + 0.08)

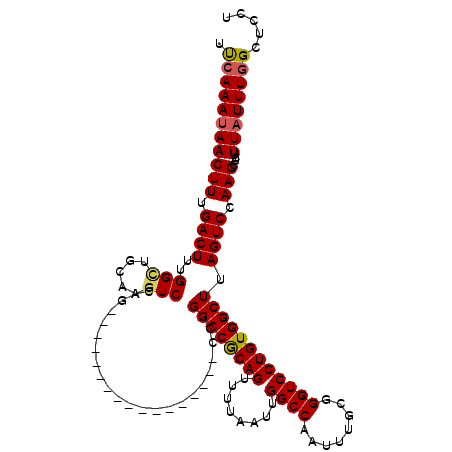

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:38 2006