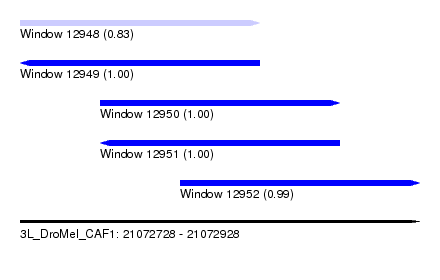
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,072,728 – 21,072,928 |
| Length | 200 |
| Max. P | 0.999950 |

| Location | 21,072,728 – 21,072,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -42.31 |
| Consensus MFE | -30.45 |
| Energy contribution | -32.21 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21072728 120 + 23771897 UCUGGAGAGCUCCACCAAACUGAAGCUAUCGCCGGACCCGAACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGCCAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAA ..((((....))))......(((((((.....((....))..((.(((((((.....))))))).))..(((.(((.(((.(((((((.....))))))).)))))).))).))))))). ( -44.10) >DroSec_CAF1 127994 120 + 1 UCUGGAGAGCUCCACCAAACUGAAGCUAUCGCCGGAGCCGAACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAA ...((..((((.((......)).))))....))(((((.(..((.(((((((.....))))))).))..(((.(((.(((.(((((((.....))))))).)))))).)))).))))).. ( -44.00) >DroSim_CAF1 34232 120 + 1 UCUGGAGAGCUCCACCAAGCUGAAGCUAUCGCCGGAGCCGAACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAA ..(((((..((.(((((.(((((.((....)).((((.....)))).......))))).))))).))..(((.(((.(((.(((((((.....))))))).)))))).)))...))))). ( -45.60) >DroEre_CAF1 50607 120 + 1 UUUGGAGAGCUCUACCAAGCGGAAAGUGUCGCCGGAGCCGAACUUCAUCAUGAUCAGAGUGGUGUAGAAAGCUACCAUGCUCAAGGCCAUCAGAGCCUUGACCAGGUAGUAGAGCUUCAG .((((((....)).))))((((......)))).((((((...((.((((((.......)))))).))...((((((.((.((((((((....).))))))).)))))))).).))))).. ( -41.10) >DroYak_CAF1 22079 120 + 1 UCUGGAGAGCUCUACCAAACUAAAUUCCUCGGUGGAGCUGAACUUCAUCAUGAUCAGAGCGGUGUAAAAUGCCACCAUGCCCAAGGCCAUCAGAGCCUUGACCAGGUAGUACAGCUUCAA ..((((((((((((((..............)))))))))...))))).........((((((((........)))).(((((((((((....).))))))....)))).....))))... ( -36.74) >consensus UCUGGAGAGCUCCACCAAACUGAAGCUAUCGCCGGAGCCGAACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAA ..(((((.(((((....................)))))....((.(((((((.....))))))).))..(((.(((.(((.(((((((.....))))))).)))))).)))...))))). (-30.45 = -32.21 + 1.76)


| Location | 21,072,728 – 21,072,848 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -38.66 |
| Energy contribution | -39.46 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21072728 120 - 23771897 UUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGGCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUUCGGGUCCGGCGAUAGCUUCAGUUUGGUGGAGCUCUCCAGA ((((((((((.((((((((((((((((.....))))))))))).)))))............((((((((.(((......)))))).))))))))))))))).....((((....)))).. ( -53.70) >DroSec_CAF1 127994 120 - 1 UUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGUCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUUCGGCUCCGGCGAUAGCUUCAGUUUGGUGGAGCUCUCCAGA ((((((((((.((((((((.(((((((.....))))))).))).)))))............(((((....(((......)))....))))))))))))))).....((((....)))).. ( -48.70) >DroSim_CAF1 34232 120 - 1 UUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGUCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUUCGGCUCCGGCGAUAGCUUCAGCUUGGUGGAGCUCUCCAGA ((((((((((.((((((((.(((((((.....))))))).))).)))))............(((((....(((......)))....))))))))))))))).....((((....)))).. ( -48.60) >DroEre_CAF1 50607 120 - 1 CUGAAGCUCUACUACCUGGUCAAGGCUCUGAUGGCCUUGAGCAUGGUAGCUUUCUACACCACUCUGAUCAUGAUGAAGUUCGGCUCCGGCGACACUUUCCGCUUGGUAGAGCUCUCCAAA ..((((((((((((..(((((((((((.....))))))))((.(((.((((..((.((.((.........)).)).))...)))))))))........)))..)))))))))).)).... ( -38.80) >DroYak_CAF1 22079 120 - 1 UUGAAGCUGUACUACCUGGUCAAGGCUCUGAUGGCCUUGGGCAUGGUGGCAUUUUACACCGCUCUGAUCAUGAUGAAGUUCAGCUCCACCGAGGAAUUUAGUUUGGUAGAGCUCUCCAGA .....(((((((((((((.((((((((.....)))))))).)).))))).....))))..))((((....(((......)))((((.((((((........)))))).))))....)))) ( -37.90) >consensus UUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGGCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUUCGGCUCCGGCGAUAGCUUCAGUUUGGUGGAGCUCUCCAGA ((((((((((.((((((((((((((((.....))))))))))).)))))............(((((....(((......)))....))))))))))))))).((((.((....)))))). (-38.66 = -39.46 + 0.80)


| Location | 21,072,768 – 21,072,888 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -47.06 |
| Consensus MFE | -42.64 |
| Energy contribution | -43.88 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21072768 120 + 23771897 AACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGCCAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAUGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAC ...((((((.((.((.((.((((((.....((((((.(((.(((((((.....))))))).))).........((......)))))))).....)))))).)).)).))....)))))). ( -51.00) >DroSec_CAF1 128034 120 + 1 AACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAC ...((((((.((.((.((.((((((.....((((((.(((.(((((((.....))))))).))).........(((....))))))))).....)))))).)).)).))....)))))). ( -50.80) >DroSim_CAF1 34272 120 + 1 AACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAC ...((((((.((.((.((.((((((.....((((((.(((.(((((((.....))))))).))).........(((....))))))))).....)))))).)).)).))....)))))). ( -50.80) >DroEre_CAF1 50647 120 + 1 AACUUCAUCAUGAUCAGAGUGGUGUAGAAAGCUACCAUGCUCAAGGCCAUCAGAGCCUUGACCAGGUAGUAGAGCUUCAGAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAG ..(((((((.((.((.(..((((((.....((((((.((.((((((((....).))))))).)).........(((....))))))))).....))))))..).)).))....))))))) ( -41.90) >DroYak_CAF1 22119 120 + 1 AACUUCAUCAUGAUCAGAGCGGUGUAAAAUGCCACCAUGCCCAAGGCCAUCAGAGCCUUGACCAGGUAGUACAGCUUCAAGGCGGUGGCCAAUGACACCAGGCAGAGCACCCAGGUGAAC ...((((((...........(((((.....((((((..(((...))).......(((((((...(((......)))))))))))))))).....))))).((........)).)))))). ( -40.80) >consensus AACUUCAUCAUGAUCAGCGUGGUGUAGAAUGCCACCAUGGACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAC ...((((((.((.((.((.((((((.....((((((.(((.(((((((.....))))))).))).........((......)))))))).....)))))).)).)).))....)))))). (-42.64 = -43.88 + 1.24)

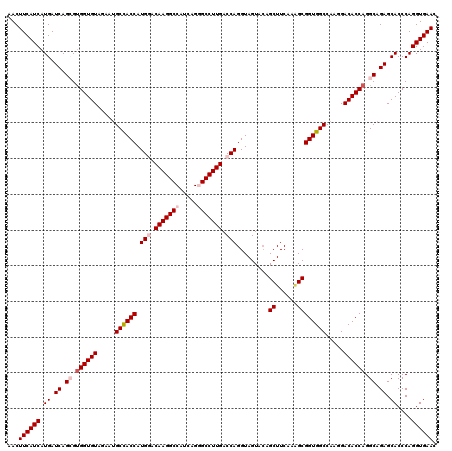
| Location | 21,072,768 – 21,072,888 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -41.40 |
| Energy contribution | -42.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21072768 120 - 23771897 GUUCACCUGGGUGCUCUGCCUGGUGUCCUUGGCCACCGCAUUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGGCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUU .((((.(...(((.((.((.((((((.....((((((((......)).........(((((((((((.....))))))))))).)))))).....)))))).)).)).)))).))))... ( -49.90) >DroSec_CAF1 128034 120 - 1 GUUCACCUGGGUGCUCUGCCUGGUGUCCUUGGCCACCGCUUUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGUCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUU .((((.(...(((.((.((.((((((.....(((((((((....))).........(((.(((((((.....))))))).))).)))))).....)))))).)).)).)))).))))... ( -47.30) >DroSim_CAF1 34272 120 - 1 GUUCACCUGGGUGCUCUGCCUGGUGUCCUUGGCCACCGCUUUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGUCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUU .((((.(...(((.((.((.((((((.....(((((((((....))).........(((.(((((((.....))))))).))).)))))).....)))))).)).)).)))).))))... ( -47.30) >DroEre_CAF1 50647 120 - 1 CUUCACCUGGGUGCUCUGCCUGGUGUCCUUGGCCACCGCUCUGAAGCUCUACUACCUGGUCAAGGCUCUGAUGGCCUUGAGCAUGGUAGCUUUCUACACCACUCUGAUCAUGAUGAAGUU (((((...((((.....))))(((((....(((....(((....))).....((((((.((((((((.....)))))))).)).)))))))....))))).............))))).. ( -38.50) >DroYak_CAF1 22119 120 - 1 GUUCACCUGGGUGCUCUGCCUGGUGUCAUUGGCCACCGCCUUGAAGCUGUACUACCUGGUCAAGGCUCUGAUGGCCUUGGGCAUGGUGGCAUUUUACACCGCUCUGAUCAUGAUGAAGUU ...((((.((((.....))))))))(((((((((((.(((((((....(.....)....)))))))...).)))))..((((..((((........)))))))).......))))).... ( -39.50) >consensus GUUCACCUGGGUGCUCUGCCUGGUGUCCUUGGCCACCGCUUUGAAGCUGUACUACCUGGUCAAGGCCCUGAUGGCCUUGGCCAUGGUGGCAUUCUACACCACGCUGAUCAUGAUGAAGUU .((((.(...(((.((.((.((((((.....((((((((......)).........(((((((((((.....))))))))))).)))))).....)))))).)).)).)))).))))... (-41.40 = -42.32 + 0.92)



| Location | 21,072,808 – 21,072,928 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -44.63 |
| Consensus MFE | -39.24 |
| Energy contribution | -40.28 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21072808 120 + 23771897 CCAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAUGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAACACCAAGUACUCCGGAUGGGCGCACUGCCCCUCAACAUCCG .(((((((.....)))))))....((.(((((.((......))(((((((..(....)..)))....(((....)))..))))..)))))))((((((((.....))))......)))). ( -46.20) >DroSec_CAF1 128074 120 + 1 ACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAACACCAAGUACUCCGGAUGGGCACAGUGCCCCUCAACAUCCG .(((((((.....)))))))....((.(((((.(((....)))(((((((..(....)..)))....(((....)))..))))..)))))))((((((((.....))))......)))). ( -47.30) >DroSim_CAF1 34312 120 + 1 ACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAACACCAAGUACUCCGGAUGGGCACAGUGCCCCUCAACAUCCG .(((((((.....)))))))....((.(((((.(((....)))(((((((..(....)..)))....(((....)))..))))..)))))))((((((((.....))))......)))). ( -47.30) >DroEre_CAF1 50687 120 + 1 UCAAGGCCAUCAGAGCCUUGACCAGGUAGUAGAGCUUCAGAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAAGACCAAGUACUCCGGAUGGGCGCACUCCCCCUCCACGUCCG ....(((((((...((((.....))))......(((....))))))))))..((((....((.....(((....)))....)).........(((.(((.......))).)))..)))). ( -39.70) >DroYak_CAF1 22159 120 + 1 CCAAGGCCAUCAGAGCCUUGACCAGGUAGUACAGCUUCAAGGCGGUGGCCAAUGACACCAGGCAGAGCACCCAGGUGAACACCAAGUACUCCGGAUGGGCGCACAUCCCCUCACCAUCCG .(((((((....).))))))....((.(((((.(((....)))(((((((..........)))....(((....)))..))))..)))))))((((((...............)))))). ( -42.66) >consensus ACAAGGCCAUCAGGGCCUUGACCAGGUAGUACAGCUUCAAAGCGGUGGCCAAGGACACCAGGCAGAGCACCCAGGUGAACACCAAGUACUCCGGAUGGGCGCACUGCCCCUCAACAUCCG .(((((((.....)))))))....((.(((((.((......))(((((((..........)))....(((....)))..))))..)))))))((((((((.....))))......)))). (-39.24 = -40.28 + 1.04)

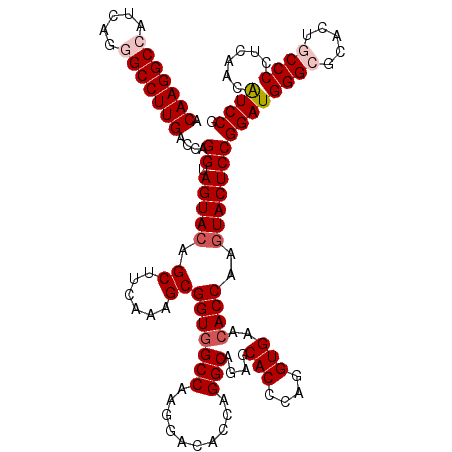
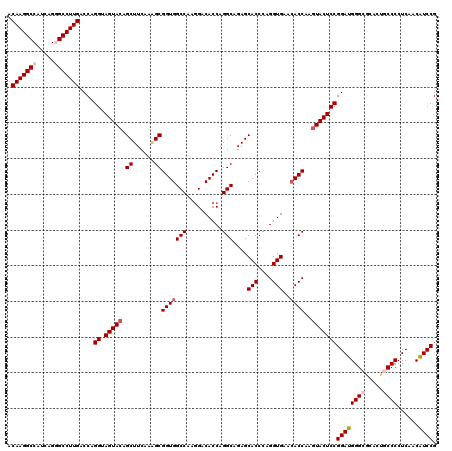
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:32 2006