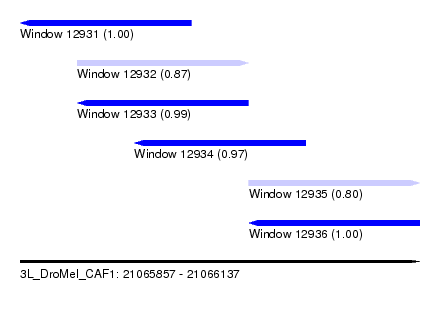
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,065,857 – 21,066,137 |
| Length | 280 |
| Max. P | 0.998520 |

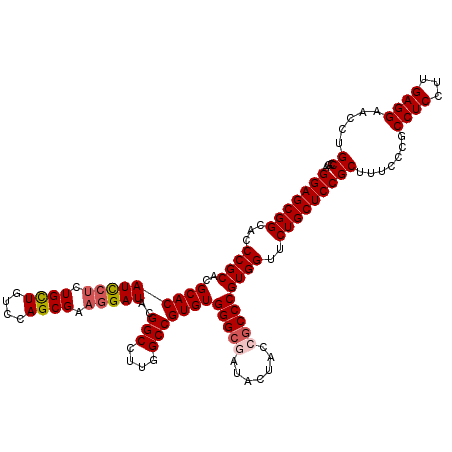
| Location | 21,065,857 – 21,065,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -50.98 |
| Consensus MFE | -49.22 |
| Energy contribution | -49.90 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21065857 120 - 23771897 GGAGCGGCACCCGCACGCACAUCCUCUGUUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUACCGCCCGUGGUUCUGCUCCGCUUUCCCGCCUCCUUGAGGAACCUGCCGA (((((((...((((..(((((((((.((((....)))).)))))..(((....)))))))(((((.......)))))))))..)))))))((.......((((...)))).....))... ( -51.60) >DroSec_CAF1 121167 120 - 1 GGAGCGGCACCCGCACGCACAUUCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCUAUACUACCCCCCGUGGUUCUGCUCCGCUUUCCCGCCUCCUUGAGGAACCUGCCGA (((((((..(((((((....(((((.((((....)))).)))))..(((....))))))))))....(((((.....))))).)))))))((.......((((...)))).....))... ( -46.00) >DroSim_CAF1 27417 120 - 1 GGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCUAUACUACCGCCCGUGGUUCUGCUCCGCUUUCCCGCCUCCUUGAGGAACCUGCCGA (((((((...((((..(((((((((.((((....)))).)))))..(((....)))))))((((.........))))))))..)))))))((.......((((...)))).....))... ( -51.00) >DroEre_CAF1 43777 120 - 1 GGAGCGGUACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUUCCGCCCGUGGUUCUGCUCCGCUUUCCCGCCUCCUUGAGGAACCUGCCGA (((((((...((((..(((((((((.((((....)))).)))))..(((....)))))))(((((.......)))))))))..)))))))((.......((((...)))).....))... ( -54.70) >DroYak_CAF1 14228 120 - 1 GGAGCGGUACCCGCACGCACAUCCGCUGCUGUCCAGCGAAAGAUACGGCCUUGGCCGUGUGGGCGAUACUUCCGCCCGUGGUUCUGCUCCGCUUUCCUGCCUCCUUGAGGAACCUGCCGA (((((((...((((.........(((((.....)))))....(((((((....)))))))(((((.......)))))))))..)))))))((.(((((.........)))))...))... ( -51.60) >consensus GGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUACCGCCCGUGGUUCUGCUCCGCUUUCCCGCCUCCUUGAGGAACCUGCCGA (((((((...((((..(((((((((.((((....)))).)))))..(((....)))))))(((((.......)))))))))..)))))))((.......((((...)))).....))... (-49.22 = -49.90 + 0.68)


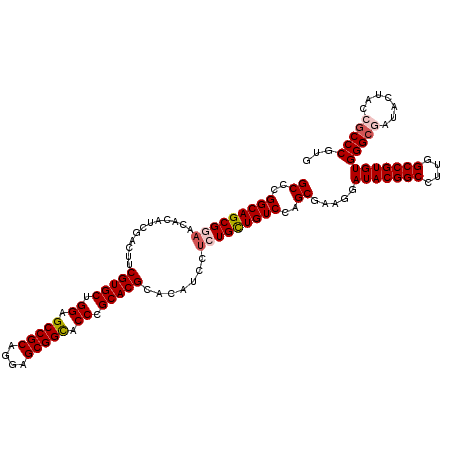
| Location | 21,065,897 – 21,066,017 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -56.60 |
| Consensus MFE | -50.48 |
| Energy contribution | -51.68 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21065897 120 + 23771897 CACGGGCGGUAGUAUCGCCCACACGGCCAAGGCCGUAUCCUUCGCUGGACAACAGAGGAUGUGCGUGCGGGUGCCGCUCCUGCGGCUCCAGCACGAAGUCGAUGUGUUCCGCUGCCGGGC ...(((((((...)))))))..(((((....))))).(((..(((.(((((.((...(((...(((((.((.(((((....))))).)).)))))..)))..))))))).)).)..))). ( -56.60) >DroSec_CAF1 121207 120 + 1 CACGGGGGGUAGUAUAGCCCACACGGCCAAGGCCGUAUCCUUCGCUGGACAGCAGAGAAUGUGCGUGCGGGUGCCGCUCCUGCGGCUCCAGCACGAAGUCGAUGUGUUCCGCUGCCGGGC ..(((.((((......))))..(((((....))))).......((.(((((.((..((.....(((((.((.(((((....))))).)).)))))...))..))))))).))..)))... ( -54.10) >DroSim_CAF1 27457 120 + 1 CACGGGCGGUAGUAUAGCCCACACGGCCAAGGCCGUAUCCUUCGCUGGACAGCAGAGGAUGUGCGUGCGGGUGCCGCUCCUGCGGCUCCAGCACGAAGUGGAUGUGUUCCGCUGCCGGGC ..(.(((((..(((((..(((((((((....)))))(((((..(((....)))..)))))...(((((.((.(((((....))))).)).)))))..)))).))))..)..))))).).. ( -61.90) >DroEre_CAF1 43817 120 + 1 CACGGGCGGAAGUAUCGCCCACACGGCCAAGGCCGUAUCCUUCGCUGGACAGCAGAGGAUGUGCGUGCGGGUACCGCUCCUGCGGCUCCAGCACGAAGUCGAUGUGUUUCGCUGCCGGGC ...((((((.....))))))..(((((....)))))........((((.((((....(((...(((((.((..((((....))))..)).)))))..)))((......)))))))))).. ( -55.20) >DroYak_CAF1 14268 120 + 1 CACGGGCGGAAGUAUCGCCCACACGGCCAAGGCCGUAUCUUUCGCUGGACAGCAGCGGAUGUGCGUGCGGGUACCGCUCCUGCGGCUCCAGCACGAAGUCGAUGUGUUUCGCUGCCGGGC ...((((((.....))))))..(((((....)))))........((((.((((....(((...(((((.((..((((....))))..)).)))))..)))((......)))))))))).. ( -55.20) >consensus CACGGGCGGUAGUAUCGCCCACACGGCCAAGGCCGUAUCCUUCGCUGGACAGCAGAGGAUGUGCGUGCGGGUGCCGCUCCUGCGGCUCCAGCACGAAGUCGAUGUGUUCCGCUGCCGGGC ..(.(((((..(...(((.((((((((....)))))((((((.(((....))).)))))))))(((((.((.(((((....))))).)).)))))........)))..)..))))).).. (-50.48 = -51.68 + 1.20)


| Location | 21,065,897 – 21,066,017 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -54.26 |
| Consensus MFE | -51.11 |
| Energy contribution | -51.91 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21065897 120 - 23771897 GCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGUUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUACCGCCCGUG ((..(((((((((.......(....)((((.((.(((((....))))).)).))))........)))))))))..)).....(((((((....)))))))(((((.......)))))... ( -55.00) >DroSec_CAF1 121207 120 - 1 GCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUUCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCUAUACUACCCCCCGUG (((((((((((((.......(....)((((.((.(((((....))))).)).))))........))))))))).........(((((((....)))))))))))................ ( -53.40) >DroSim_CAF1 27457 120 - 1 GCCCGGCAGCGGAACACAUCCACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCUAUACUACCGCCCGUG ((..(((((((((.....)))....(((((.((.(((((....))))).)).)))))..........))))))..)).....(((((((....)))))))((((.........))))... ( -54.60) >DroEre_CAF1 43817 120 - 1 GCCCGGCAGCGAAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGUACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUUCCGCCCGUG ((((((((.((((..........)))))))))).))(((.((.((((....(((.((((((((((.((((....)))).)))))..(((....)))))))).)))......))))))))) ( -53.20) >DroYak_CAF1 14268 120 - 1 GCCCGGCAGCGAAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGUACCCGCACGCACAUCCGCUGCUGUCCAGCGAAAGAUACGGCCUUGGCCGUGUGGGCGAUACUUCCGCCCGUG (..((((((((.........(....)((((.((.(((((....))))).)).)))).......))))))))..)..(....)(((((((....)))))))(((((.......)))))... ( -55.10) >consensus GCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAAGGAUACGGCCUUGGCCGUGUGGGCGAUACUACCGCCCGUG ((..(((((((((............(((((.((.(((((....))))).)).))))).......)))))))))..)).....(((((((....)))))))(((((.......)))))... (-51.11 = -51.91 + 0.80)

| Location | 21,065,937 – 21,066,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -43.84 |
| Energy contribution | -43.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21065937 120 - 23771897 CUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUUAUCGCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGUUGUCCAGCGAA ........(((.(((((.(((((((((((.((........)).))).))))))............(((((.((.(((((....))))).)).)))))...........)).)))))))). ( -45.70) >DroSec_CAF1 121247 120 - 1 CUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUGACCGCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUUCUCUGCUGUCCAGCGAA ........(((.(((((.(((((((((((.((........)).))).))))))............(((((.((.(((((....))))).)).)))))...........)).)))))))). ( -47.80) >DroSim_CAF1 27497 120 - 1 CUUCUCCAUCGUCUGGAGUGUCUGCUCAGUGUCCUUGACCGCCCGGCAGCGGAACACAUCCACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAA ...(((((.....))))).(((.((.....))....)))(((..(((((((((.....)))....(((((.((.(((((....))))).)).)))))..........))))))..))).. ( -44.70) >DroEre_CAF1 43857 120 - 1 CUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUAACCGCCCGGCAGCGAAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGUACCCGCACGCACAUCCUCUGCUGUCCAGCGAA ........(((.(((((((((((((((((((.........((((((((.((((..........)))))))))).))))).))))))).))))....(((.......)))..)))))))). ( -47.10) >DroYak_CAF1 14308 120 - 1 CUUCUCCAUCGACUGGAGGGUCUGCUCCGUGUCCUUUACCGCCCGGCAGCGAAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGUACCCGCACGCACAUCCGCUGCUGUCCAGCGAA ........(((.(((((((((((((((((((.....))).((((((((.((((..........)))))))))).))....))))))).))))(((.((......)))))..)))))))). ( -51.10) >consensus CUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUGACCGCCCGGCAGCGGAACACAUCGACUUCGUGCUGGAGCCGCAGGAGCGGCACCCGCACGCACAUCCUCUGCUGUCCAGCGAA ........(((.(((((((((((((((((((.........((((((((.((((..........)))))))))).))))).))))))).))))....(((.......)))..)))))))). (-43.84 = -43.84 + -0.00)



| Location | 21,066,017 – 21,066,137 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21066017 120 + 23771897 GAUAAAGGACACGGAGCAGACCCUCCAGACGAUGGAGAAGGAGCGCACGGAAUACCGCGCCGCUCUCUCCUGGAUGAAGUCCGCCAGCCAGGAACUGGAUCCCGAUACGGGCAAGGGUCU .................(((((((((((...........(((((((.(((....))).).)))))).((((((.((........)).)))))).))))).((((...))))...)))))) ( -44.80) >DroSec_CAF1 121327 120 + 1 GGUCAAGGACACGGAGCAGACCCUCCAGACGAUGGAGAAGGAGCGCACGGAAUACCGCGCCGCUCUCUCCUGGAUGAAGUCAGCCAGCCAGGAGCUGGAUCCCGAUACGGGCAAGGGUCU .(((...))).......(((((((((((...........(((((((.(((....))).).))))))(((((((.((........)).)))))))))))).((((...))))...)))))) ( -47.30) >DroSim_CAF1 27577 120 + 1 GGUCAAGGACACUGAGCAGACACUCCAGACGAUGGAGAAGGAGCGCACGGAAUACCGCGCCGCUCUCUCCUGGAUGAAGUCAGCCAGCCAGGAGCUGGAUCCCGAUACGGGCAAAGGUCU ......((((.((...(((...(((((.....)))))..(((((((.(((....))).).))))))(((((((.((........)).))))))))))...((((...))))...)))))) ( -43.00) >DroEre_CAF1 43937 120 + 1 GGUUAAGGACACGGAGCAGACCCUCCAGACGAUGGAGAAGGAGCGCACGGAAUACCGCGCCGCUCUCUCGUGGAUGAAGUCCGCAAGCCAGGAACUGGAUCCCGACACGGGCAAGGGUCU .(((..(....)..)))(((((((((((....(((.((..((((((.(((....))).).)))))..))((((((...))))))...)))....))))).((((...))))...)))))) ( -49.00) >DroYak_CAF1 14388 120 + 1 GGUAAAGGACACGGAGCAGACCCUCCAGUCGAUGGAGAAGGAGCGCACGGAAUAUCGUGCCGCUCUCUCGUGGAUGAAGUCCGCAAGCCAGGAACUGGAUCCCGAUACGGGAAAGGGUCU .................((((((((((((...(((.((..(((((((((......)))).)))))..))((((((...))))))...)))...))))))(((((...)))))..)))))) ( -52.60) >consensus GGUAAAGGACACGGAGCAGACCCUCCAGACGAUGGAGAAGGAGCGCACGGAAUACCGCGCCGCUCUCUCCUGGAUGAAGUCCGCCAGCCAGGAACUGGAUCCCGAUACGGGCAAGGGUCU .................(((((((((((....(((.((..((((((.(((....))).).)))))..)).(((((...)))))....)))....))))).((((...))))...)))))) (-38.08 = -38.72 + 0.64)



| Location | 21,066,017 – 21,066,137 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -49.36 |
| Consensus MFE | -43.66 |
| Energy contribution | -44.30 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21066017 120 - 23771897 AGACCCUUGCCCGUAUCGGGAUCCAGUUCCUGGCUGGCGGACUUCAUCCAGGAGAGAGCGGCGCGGUAUUCCGUGCGCUCCUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUUAUC (((((((((((.((..(((((......))))))).)))(((.....))).((((((((((.(((((....))))))))))..)))))........))))))))................. ( -51.00) >DroSec_CAF1 121327 120 - 1 AGACCCUUGCCCGUAUCGGGAUCCAGCUCCUGGCUGGCUGACUUCAUCCAGGAGAGAGCGGCGCGGUAUUCCGUGCGCUCCUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUGACC ((((((...((((...)))).((((((((((((.(((......))).))))))).(((((.(((((....))))))))))............)))))))))))................. ( -51.60) >DroSim_CAF1 27577 120 - 1 AGACCUUUGCCCGUAUCGGGAUCCAGCUCCUGGCUGGCUGACUUCAUCCAGGAGAGAGCGGCGCGGUAUUCCGUGCGCUCCUUCUCCAUCGUCUGGAGUGUCUGCUCAGUGUCCUUGACC ((((.....((((...)))).....(((((.((((((.((....)).)))((((((((((.(((((....))))))))))..)))))...))).)))))))))................. ( -46.50) >DroEre_CAF1 43937 120 - 1 AGACCCUUGCCCGUGUCGGGAUCCAGUUCCUGGCUUGCGGACUUCAUCCACGAGAGAGCGGCGCGGUAUUCCGUGCGCUCCUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUAACC ((((((((..(((((((((((......)))))))..))))...........(((((((((.(((((....))))))))))..)))).........))))))))................. ( -48.20) >DroYak_CAF1 14388 120 - 1 AGACCCUUUCCCGUAUCGGGAUCCAGUUCCUGGCUUGCGGACUUCAUCCACGAGAGAGCGGCACGAUAUUCCGUGCGCUCCUUCUCCAUCGACUGGAGGGUCUGCUCCGUGUCCUUUACC ((((((..(((((...)))))(((((((..(((((((.(((.....))).)))).(((((.((((......))))))))).....)))..)))))))))))))................. ( -49.50) >consensus AGACCCUUGCCCGUAUCGGGAUCCAGUUCCUGGCUGGCGGACUUCAUCCAGGAGAGAGCGGCGCGGUAUUCCGUGCGCUCCUUCUCCAUCGUCUGGAGGGUCUGCUCCGUGUCCUUGACC ((((((...((((...)))).((((((((((((.(((......))).))))))).(((((.(((((....))))))))))............)))))))))))................. (-43.66 = -44.30 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:16 2006