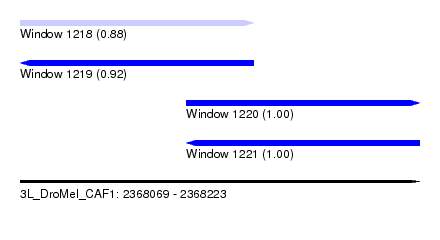
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,368,069 – 2,368,223 |
| Length | 154 |
| Max. P | 0.999917 |

| Location | 2,368,069 – 2,368,159 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
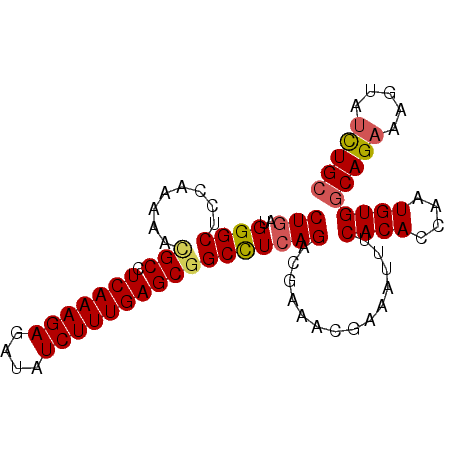
>3L_DroMel_CAF1 2368069 90 + 23771897 CUGAUGGGCUCCAAAAACGCCUCAAAGACAUAUCUUUGAGCUGCCUCAGACGAAACAAAAUUUCACACCAAUGUGGCAGAAAGUAUCUGC ((((.((((.........))((((((((....))))))))...))))))..............((((....))))(((((.....))))) ( -20.50) >DroSec_CAF1 116752 90 + 1 CUGUUGGGCUCCAAAAACGCCUCAAAGAGAUAUCUUUGAGCGGCCUCAGACGAAACGAAAUUUCACACCAAUGUGGCAGAAAGUAUCUGC (((..((((........(((.(((((((....)))))))))))))))))..............((((....))))(((((.....))))) ( -23.50) >DroSim_CAF1 117812 90 + 1 CUGUUGGGCUCCAAAAACGCCUCAAAGAGAUAUCUUUGAGCGGCCUCAGACGAAACGAAAUUUCACACCAAUGUGGCAGAAAGUAUCUGC (((..((((........(((.(((((((....)))))))))))))))))..............((((....))))(((((.....))))) ( -23.50) >DroEre_CAF1 124063 83 + 1 CUAAUGGGCUCCAAAAAUGCCUCAAAGAGAUAUCUUUGAGCGGCCUCAGACAAAAUU--AUUUCACACCAAUGUGGCAGAA-----UUGG ....((((((........((.(((((((....))))))))))))).)).........--........(((((........)-----)))) ( -17.50) >DroYak_CAF1 121207 88 + 1 CUGAUGGGCUCCAAAAACGCCUCAAAGAGAUAUCUUUGAGCGGCUUCAGACAUAACG--AUUUCACACCAAUGUGGCAGAAAGUAUCUGC (((((((...)))....(((.(((((((....))))))))))...))))........--....((((....))))(((((.....))))) ( -22.90) >consensus CUGAUGGGCUCCAAAAACGCCUCAAAGAGAUAUCUUUGAGCGGCCUCAGACGAAACGAAAUUUCACACCAAUGUGGCAGAAAGUAUCUGC (((..((((........(((.(((((((....)))))))))))))))))..............((((....))))(((((.....))))) (-19.36 = -19.72 + 0.36)

| Location | 2,368,069 – 2,368,159 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2368069 90 - 23771897 GCAGAUACUUUCUGCCACAUUGGUGUGAAAUUUUGUUUCGUCUGAGGCAGCUCAAAGAUAUGUCUUUGAGGCGUUUUUGGAGCCCAUCAG (((((.....)))))((((....)))).......((((((...((.((..((((((((....)))))))))).))..))))))....... ( -25.70) >DroSec_CAF1 116752 90 - 1 GCAGAUACUUUCUGCCACAUUGGUGUGAAAUUUCGUUUCGUCUGAGGCCGCUCAAAGAUAUCUCUUUGAGGCGUUUUUGGAGCCCAACAG (((((.....)))))....((((.(((((((...))))).((..((..((((((((((....))))))).)))..))..))))))))... ( -28.00) >DroSim_CAF1 117812 90 - 1 GCAGAUACUUUCUGCCACAUUGGUGUGAAAUUUCGUUUCGUCUGAGGCCGCUCAAAGAUAUCUCUUUGAGGCGUUUUUGGAGCCCAACAG (((((.....)))))....((((.(((((((...))))).((..((..((((((((((....))))))).)))..))..))))))))... ( -28.00) >DroEre_CAF1 124063 83 - 1 CCAA-----UUCUGCCACAUUGGUGUGAAAU--AAUUUUGUCUGAGGCCGCUCAAAGAUAUCUCUUUGAGGCAUUUUUGGAGCCCAUUAG ((((-----...((((.....(((.(.(.((--(....))).).).)))..(((((((....)))))))))))...)))).......... ( -20.40) >DroYak_CAF1 121207 88 - 1 GCAGAUACUUUCUGCCACAUUGGUGUGAAAU--CGUUAUGUCUGAAGCCGCUCAAAGAUAUCUCUUUGAGGCGUUUUUGGAGCCCAUCAG (((((.....)))))((((....))))....--.......((..(((.((((((((((....))))))).))).)))..))......... ( -25.50) >consensus GCAGAUACUUUCUGCCACAUUGGUGUGAAAUUUCGUUUCGUCUGAGGCCGCUCAAAGAUAUCUCUUUGAGGCGUUUUUGGAGCCCAUCAG (((((.....))))).....(((.(((((((...))))).((..(((.((((((((((....))))))).))).)))..))))))).... (-21.86 = -22.50 + 0.64)

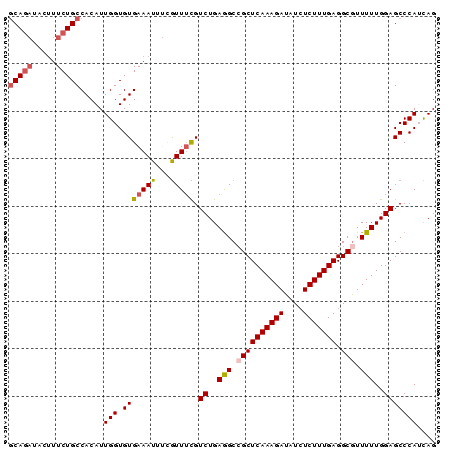
| Location | 2,368,133 – 2,368,223 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
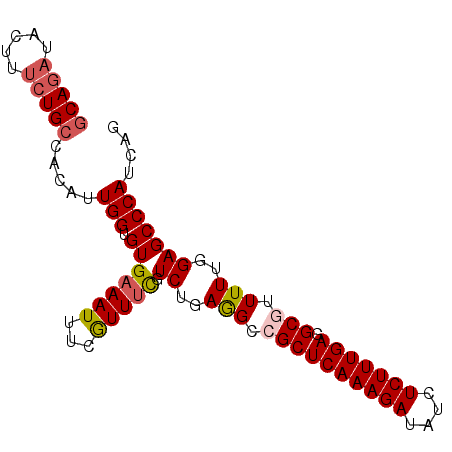
>3L_DroMel_CAF1 2368133 90 + 23771897 ACACCAAUGUGGCAGAAAGUAUCUGCUGAAUUGGUGGAUGCCCGCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAAAGAUUG .((((.....((((((.....))))))((.((((((((((..(((............)))...))))))))))))))))........... ( -29.40) >DroSec_CAF1 116816 90 + 1 ACACCAAUGUGGCAGAAAGUAUCUGCUGGAUUGGUGGCUGCCCCCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAAAGAUUG .(((((((.(((((((.....))))))).))))))).........(((.((......)))))...((((((....))))))......... ( -29.40) >DroSim_CAF1 117876 90 + 1 ACACCAAUGUGGCAGAAAGUAUCUGCUGGAUUGGUGGCUGCCCCCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAAAGAUUG .(((((((.(((((((.....))))))).))))))).........(((.((......)))))...((((((....))))))......... ( -29.40) >DroEre_CAF1 124125 85 + 1 ACACCAAUGUGGCAGAA-----UUGGUGGAUUGGUGGAUACCCCCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAGAGAUUG .(((((((........)-----))))))..((((.((....))))))........((((.((...((((((....))))))..)).)))) ( -25.30) >DroYak_CAF1 121269 90 + 1 ACACCAAUGUGGCAGAAAGUAUCUGCUGGAUUGGUGAAUGCCCCCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAAAGAUUG .(((((((.(((((((.....))))))).))))))).........(((.((......)))))...((((((....))))))......... ( -29.20) >consensus ACACCAAUGUGGCAGAAAGUAUCUGCUGGAUUGGUGGAUGCCCCCGACUACCUGACAGUGUCCCAUCCAUCAAUCGGUGGAAAAAGAUUG .(((((((.(((((((.....))))))).))))))).........(((.((......)))))...((((((....))))))......... (-26.76 = -27.00 + 0.24)

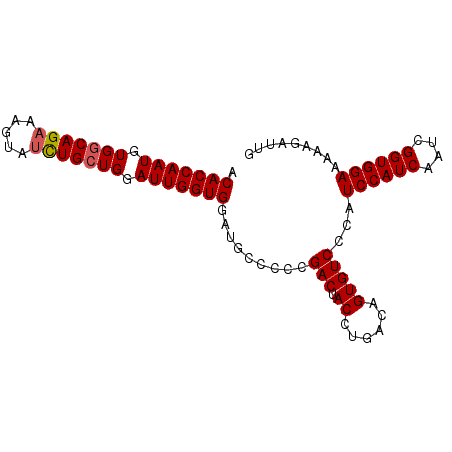
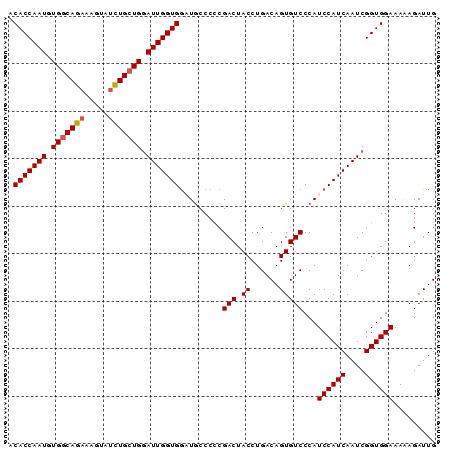
| Location | 2,368,133 – 2,368,223 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.54 |
| SVM RNA-class probability | 0.999917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2368133 90 - 23771897 CAAUCUUUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGCGGGCAUCCACCAAUUCAGCAGAUACUUUCUGCCACAUUGGUGU ..........((..((((((...(((((....)))))...))))))..))....(((((((...(((((.....)))))...))))))). ( -29.50) >DroSec_CAF1 116816 90 - 1 CAAUCUUUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGGGGGCAGCCACCAAUCCAGCAGAUACUUUCUGCCACAUUGGUGU .........(((.(((((((...(((((....)))))...))))))))))....(((((((...(((((.....)))))...))))))). ( -32.90) >DroSim_CAF1 117876 90 - 1 CAAUCUUUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGGGGGCAGCCACCAAUCCAGCAGAUACUUUCUGCCACAUUGGUGU .........(((.(((((((...(((((....)))))...))))))))))....(((((((...(((((.....)))))...))))))). ( -32.90) >DroEre_CAF1 124125 85 - 1 CAAUCUCUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGGGGGUAUCCACCAAUCCACCAA-----UUCUGCCACAUUGGUGU .........(((.(((((((...(((((....)))))...))))))))))............((((((-----(........))))))). ( -24.70) >DroYak_CAF1 121269 90 - 1 CAAUCUUUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGGGGGCAUUCACCAAUCCAGCAGAUACUUUCUGCCACAUUGGUGU .........(((.(((((((...(((((....)))))...))))))))))....(((((((...(((((.....)))))...))))))). ( -32.70) >consensus CAAUCUUUUUCCACCGAUUGAUGGAUGGGACACUGUCAGGUAGUCGGGGGCAUCCACCAAUCCAGCAGAUACUUUCUGCCACAUUGGUGU .........(((.(((((((...(((((....)))))...))))))))))....(((((((...(((((.....)))))...))))))). (-28.78 = -29.58 + 0.80)


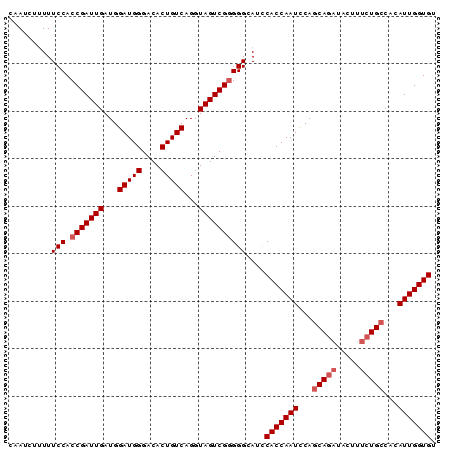
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:20 2006