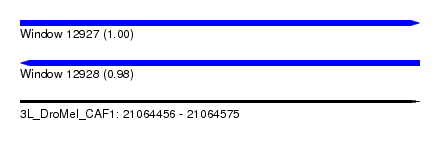
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,064,456 – 21,064,575 |
| Length | 119 |
| Max. P | 0.996122 |

| Location | 21,064,456 – 21,064,575 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21064456 119 + 23771897 CAAAACAUGCACACACCCACAUGCAGGUGGCAAAAACUCCACCUCACAACCCGUUU-UUCUCAUUGCCAAGAAAACUACUGAGCUUUGCACAGACUUCUUCACUUUCUUCUUGUGCGUGC .....((((((((........((.((((((........))))))))........((-((((........))))))...(((.((...)).)))..................)))))))). ( -24.80) >DroSec_CAF1 119465 120 + 1 CAAAACAUGCACACACCCACAUGCAGGUGGCGAAAACUCCACCUCACAACCCGUUUUUUCUCAUUGCCAAGAAAACUACUGAGCUUUGCACACACUUCUUCACUUUCUUCUUGUGCGUGC .....((((((((........((.((((((.(....).))))))))......((.((((((........))))))..))................................)))))))). ( -23.40) >DroSim_CAF1 25723 120 + 1 CAAAACAUGCACACACCCACAUGCAGGUGGCGAAAACUCCACCUCACAACCCGUUUUUUCUCAUUGCCAAGAAAACUACUGAGCUUUGCACACACUUCUUCACUUUCUUCUUGUGCGUGC .....((((((((........((.((((((.(....).))))))))......((.((((((........))))))..))................................)))))))). ( -23.40) >DroEre_CAF1 42312 120 + 1 CAAAACAUGCACACACCCACAUGCAGGUGGCAAAAACUCCACCUCACAACCCGUUUUUUCUUAUUGCGAAGAAAACUACUGAGCUUUGCACACACUUCUUCACUUUCUUCUUGUGCGUGA .....((((((((........((.((((((........))))))))..................(((((((............))))))).....................)))))))). ( -26.80) >DroYak_CAF1 12732 120 + 1 GGAAACAUGCUCACACCCACAUGCAGGUGGCAAAAACUCCACCUCACAACCCGUUUUUUCUCAUUGCCAAGAAAACUACUGAGCUUUGCACACACUUCUUCACUUUCUUCUUGUGCGUGC (....)..(((((........((.((((((........))))))))......((.((((((........))))))..)))))))...((((.(((.................))).)))) ( -27.13) >consensus CAAAACAUGCACACACCCACAUGCAGGUGGCAAAAACUCCACCUCACAACCCGUUUUUUCUCAUUGCCAAGAAAACUACUGAGCUUUGCACACACUUCUUCACUUUCUUCUUGUGCGUGC .....((((((((........((.((((((........))))))))...............((..((..((.......))..))..)).......................)))))))). (-21.54 = -21.58 + 0.04)

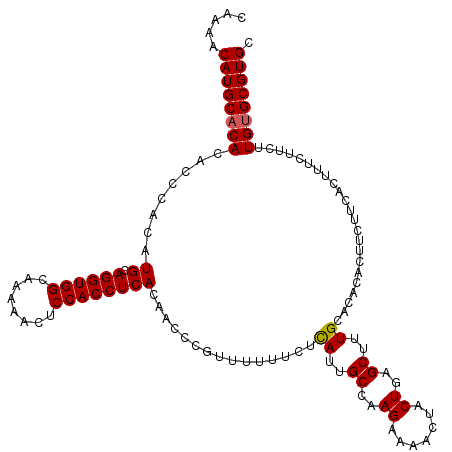
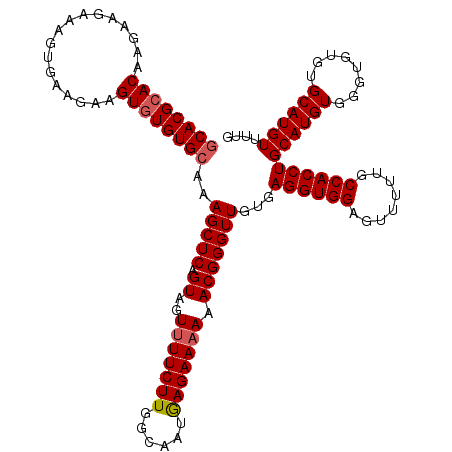
| Location | 21,064,456 – 21,064,575 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -32.19 |
| Energy contribution | -32.63 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21064456 119 - 23771897 GCACGCACAAGAAGAAAGUGAAGAAGUCUGUGCAAAGCUCAGUAGUUUUCUUGGCAAUGAGAA-AAACGGGUUGUGAGGUGGAGUUUUUGCCACCUGCAUGUGGGUGUGUGCAUGUUUUG ((((((((.........((.((((((.(((.((...)).)))....)))))).))........-...((.((.((.((((((........)))))))))).)).))))))))........ ( -37.70) >DroSec_CAF1 119465 120 - 1 GCACGCACAAGAAGAAAGUGAAGAAGUGUGUGCAAAGCUCAGUAGUUUUCUUGGCAAUGAGAAAAAACGGGUUGUGAGGUGGAGUUUUCGCCACCUGCAUGUGGGUGUGUGCAUGUUUUG ((((((((.................))))))))..(((((.((..(((((((......))))))).)))))))...((((((.(....).))))))((((((........)))))).... ( -35.53) >DroSim_CAF1 25723 120 - 1 GCACGCACAAGAAGAAAGUGAAGAAGUGUGUGCAAAGCUCAGUAGUUUUCUUGGCAAUGAGAAAAAACGGGUUGUGAGGUGGAGUUUUCGCCACCUGCAUGUGGGUGUGUGCAUGUUUUG ((((((((.................))))))))..(((((.((..(((((((......))))))).)))))))...((((((.(....).))))))((((((........)))))).... ( -35.53) >DroEre_CAF1 42312 120 - 1 UCACGCACAAGAAGAAAGUGAAGAAGUGUGUGCAAAGCUCAGUAGUUUUCUUCGCAAUAAGAAAAAACGGGUUGUGAGGUGGAGUUUUUGCCACCUGCAUGUGGGUGUGUGCAUGUUUUG .(((((((.........(((....((.(((.(((((((((.........(((((((((..(......)..)))))))))..))).))))))))))).)))....)))))))......... ( -32.72) >DroYak_CAF1 12732 120 - 1 GCACGCACAAGAAGAAAGUGAAGAAGUGUGUGCAAAGCUCAGUAGUUUUCUUGGCAAUGAGAAAAAACGGGUUGUGAGGUGGAGUUUUUGCCACCUGCAUGUGGGUGUGAGCAUGUUUCC ((((((((.................))))))))...(((((....(((((((......)))))))..((.((.((.((((((........)))))))))).))....)))))........ ( -35.83) >consensus GCACGCACAAGAAGAAAGUGAAGAAGUGUGUGCAAAGCUCAGUAGUUUUCUUGGCAAUGAGAAAAAACGGGUUGUGAGGUGGAGUUUUUGCCACCUGCAUGUGGGUGUGUGCAUGUUUUG ((((((((.................))))))))..(((((.((..(((((((......))))))).)))))))...((((((........))))))((((((........)))))).... (-32.19 = -32.63 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:07 2006