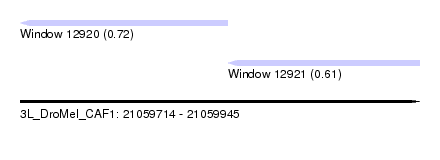
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,059,714 – 21,059,945 |
| Length | 231 |
| Max. P | 0.719436 |

| Location | 21,059,714 – 21,059,834 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -27.82 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21059714 120 - 23771897 CACCAAUACCACUUUAAAGUACCAGCCUCGAAAUCAGGUGCGCUUUUAGAAGCCUCCUACAUGGAAGUUUCCAACUCGAGCUGCGCCUGAUCGACUGACCCACAAAAAGCCUUUGGGCGG ........((............(((..((((..((((((((((((((.(((((.(((.....))).))))).))...)))).)))))))))))))))...........(((....))))) ( -37.30) >DroSec_CAF1 114716 120 - 1 CACCAAUACCACUUAAAAGUACCAGCCUCAAAUUCAGAUGCGCUUUUAGAAGCCUCCUACGUGGAAGUUUCCAACUCGAGUUGCGCCUGAUCGACUGACCCACAAAAAGCCUUUGGGCGG ........((............(((..((....((((..((((.....(((((.(((.....))).))))).(((....)))))))))))..)))))...........(((....))))) ( -25.40) >DroSim_CAF1 20985 120 - 1 CACCAAUACCACUUAAAAGUACCAGCCUCGAAUUCAGAUGCGCUUUUAGAAGCCUCCUACAUGGAAGUUUCCAACUCGAGUUGCGCCUGAUCGACUGACCCACAAAAAGCCUUUGGGCGG ........((............(((..((((..((((..((((.....(((((.(((.....))).))))).(((....))))))))))))))))))...........(((....))))) ( -29.40) >DroEre_CAF1 37251 116 - 1 CACCAAUACCACUUAAAAA---CAGCCUCGAAUUCAGAUGCGCUUUUAGAAGCCUCCUACAUGGAAGUUUCCAACUCGAGUUGCGCCUGAUCGACUGACCCACAA-AAGCCUUUGGGCGG ........((.........---(((..((((..((((..((((.....(((((.(((.....))).))))).(((....))))))))))))))))))........-..(((....))))) ( -29.60) >DroYak_CAF1 8117 117 - 1 ---CAAUACCACUUAAAAAUACCAGCCUCGAAUUCAGAUGCGCUUUUAGAAGCCUCCUACAUGGAAGUUUCCAACUCGAGUUGCGCCUGAUCGACUGACCCACAAAAAGCCUUUGGGCGG ---.....((............(((..((((..((((..((((.....(((((.(((.....))).))))).(((....))))))))))))))))))...........(((....))))) ( -29.40) >consensus CACCAAUACCACUUAAAAGUACCAGCCUCGAAUUCAGAUGCGCUUUUAGAAGCCUCCUACAUGGAAGUUUCCAACUCGAGUUGCGCCUGAUCGACUGACCCACAAAAAGCCUUUGGGCGG ........((............(((..((((..((((.(((((((((.(((((.(((.....))).))))).))...)))).))).)))))))))))...........(((....))))) (-27.82 = -27.86 + 0.04)


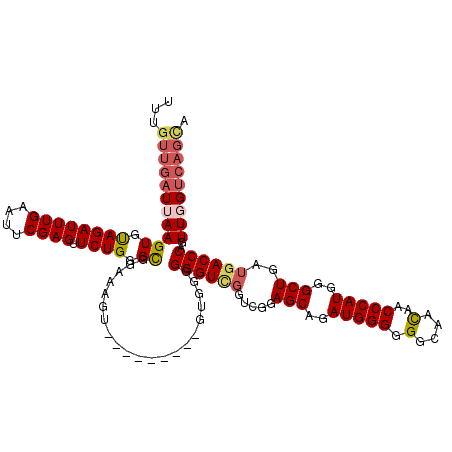
| Location | 21,059,834 – 21,059,945 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -26.52 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21059834 111 - 23771897 UUUGUUGAUUAAGUUCAGAUUUGAAUUCGAGUUCUGGGCGGUAAGU---------GUGGUGGGUCGGCCGAAGCAGAUGGGGGGCAAUAACCCAUGGGCUGAUGACCCAGUUGGUCAGCA ..(((((((((((((((((((((....)))).))))))).......---------....(((((((.....(((..(((((.........)))))..)))..))))))).)))))))))) ( -42.70) >DroSec_CAF1 114836 107 - 1 UUUGUUGAUUAAGUGUAGAUUUGAAUUCGAGUUCUGGGCGGAAAGU---------GUGGGGGGU----CGGAGCAGAUGGGGGGCAACAACCCAUGGGCUGAUGACCCAGUUGGUCAGUA ....((((((((((.((((((((....)))).)))).)).......---------.....((((----((.(((..(((((.(....)..)))))..)))..))))))..)))))))).. ( -37.30) >DroSim_CAF1 21105 111 - 1 UUUGUUGAUUAAGUGUAGAUUUGAAUUCGAGUUCUGGGCGGAAAGU---------GUGGGGGGUCGGUCGGAGCAGAUGGGGGGCAACAACCCAUGGGCUGAUGACCCAGUUGGUCAGUA ....((((((((((.((((((((....)))).)))).)).......---------.....((((((.((..(((..(((((.(....)..)))))..)))))))))))..)))))))).. ( -36.30) >DroEre_CAF1 37367 110 - 1 UUUGUUGAUAAAGUGUAGAUUUGAAUUCGAGUUCUGGGUGGGAAGU---------GUGUGGGGUUGGUCGGAGCAGAUGGG-GGCAACAACCCAUGGGCUGAUGACCCAAUUGGCCAGCC ...((..((........((((((....)))))).(((((.(..(((---------.((((((.((.(((......))).))-(....)..)))))).)))..).)))))))..))..... ( -31.80) >DroYak_CAF1 8234 114 - 1 UUUCUUGAUAAAGUGUAGAUUUGACUUCGAGUUCUGGGUGGGAAGUGGGAAGUGUGUGUGGGGUUGGUCGGAGCAGAUGGG-GGCAACAACCCAUGGGCUGAUGACCCAAUUGGU----- ....(..((.((.(.((.((...(((((.(.((((.....)))).).).))))..)).)).).))(((((.(((..(((((-........)))))..)))..)))))..))..).----- ( -26.90) >consensus UUUGUUGAUUAAGUGUAGAUUUGAAUUCGAGUUCUGGGCGGAAAGU_________GUGGGGGGUCGGUCGGAGCAGAUGGGGGGCAACAACCCAUGGGCUGAUGACCCAGUUGGUCAGCA ...(((((((((((.((((((((....)))).)))).)).....................((((((.....(((..(((((.(....)..)))))..)))..))))))..))))))))). (-26.52 = -27.44 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:00 2006