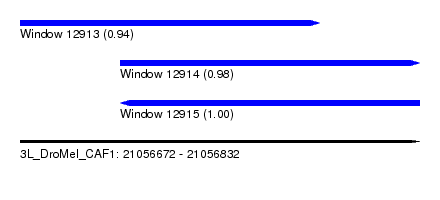
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,056,672 – 21,056,832 |
| Length | 160 |
| Max. P | 0.996644 |

| Location | 21,056,672 – 21,056,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -34.19 |
| Consensus MFE | -30.04 |
| Energy contribution | -30.56 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056672 120 + 23771897 AGACGCUGCGUCAUCGGAUGGAAGAUCUGUUCAAAGUCCAAAGCGCCUUACUAUAAAUAGGCAAACAAUUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAG .((((...)))).((((((((.....))))))((((((......((((..........)))).......(((((((((((((.(((.....))).))))))).))))))))))))..)). ( -31.90) >DroSec_CAF1 111723 120 + 1 AGACGCUGCGUCAUCGGAUCGAAGAUCUGUUCAAAGUCCACAGCGCCUGACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAG ....((((.(....((((((...)))))).........).))))(((((........)))))......((((((((((((((.(((.....))).))))))).))))))).......... ( -34.02) >DroSim_CAF1 17982 120 + 1 AGACGCUGCGUCAUCGGAUCGAAGAUCUGUUCAAAGUCCACAGCGCCUAACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAG ....((((.(....((((((...)))))).........).))))(((((........)))))......((((((((((((((.(((.....))).))))))).))))))).......... ( -34.82) >DroEre_CAF1 34365 120 + 1 AGACGCUGCGUCAUCGGAUAGAAGAUCUGUUCAAAGUCCAAAGCGCCCAGCUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUCAGCG ...(((((.(((...((((((.....)))))).........(((.....))).......)))......((((((((((((((.(((.....))).))))))).))))))).....))))) ( -37.00) >DroYak_CAF1 5173 119 + 1 AGACGCUGCGUCAUCGGAUCGAAGAUCUGUUCAAAGUCCAAAGCGCCUAGCUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGACUCU-UGCAGAUUUGUAGAUUUUCAGAG .((((...))))...(((((..((((((((....((((....(((((((........)))))......((((.....))))......)).))))..-.))))))))...)))))...... ( -33.20) >consensus AGACGCUGCGUCAUCGGAUCGAAGAUCUGUUCAAAGUCCAAAGCGCCUAACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAG .((((...))))...((((.(((......)))...)))).....(((((........)))))......((((((((((((((.(((.....))).))))))).))))))).......... (-30.04 = -30.56 + 0.52)

| Location | 21,056,712 – 21,056,832 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -32.60 |
| Energy contribution | -33.12 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056712 120 + 23771897 AAGCGCCUUACUAUAAAUAGGCAAACAAUUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU ....(((...((((((((..((((....))))...(((((((.(((.....))).))))))))))))))).......(((....)))......))).(((((.((....))..))))).. ( -33.00) >DroSec_CAF1 111763 120 + 1 CAGCGCCUGACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU .((((((((........)))))......((((((((((((((.(((.....))).))))))).))))))).......(((....))).......)))(((((.((....))..))))).. ( -35.50) >DroSim_CAF1 18022 120 + 1 CAGCGCCUAACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU .((((((((........)))))......((((((((((((((.(((.....))).))))))).))))))).......(((....))).......)))(((((.((....))..))))).. ( -36.30) >DroEre_CAF1 34405 120 + 1 AAGCGCCCAGCUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUCAGCGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU .(((...(((((.......(((......((((.....))))......)))((((((((((.((...........)).))))))))))......)))))...)))..((((......)))) ( -38.80) >DroYak_CAF1 5213 119 + 1 AAGCGCCUAGCUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGACUCU-UGCAGAUUUGUAGAUUUUCAGAGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU .((((((((........)))))......((((((((((((((...........)).-))))).)))))))....((.(((....))).))....)))(((((.((....))..))))).. ( -33.40) >consensus AAGCGCCUAACUAUAAAUAGGCAAACAACUGCAGACUGCAGAAACUGGCCGAGUCUUUGCAGAUUUGUAGAUUUUAAGAGAAGACUCCGAUCCGGCUGCCAGCUGAAAGCAAUCUGGCUU .((((((((........)))))......((((((((((((((.(((.....))).))))))).)))))))........................)))(((((.((....))..))))).. (-32.60 = -33.12 + 0.52)



| Location | 21,056,712 – 21,056,832 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -34.67 |
| Energy contribution | -35.31 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056712 120 - 23771897 AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCUCUUAAAAUCUACAAAUCUGCAAAGACUCGGCCAGUUUCUGCAGUCUGCAAUUGUUUGCCUAUUUAUAGUAAGGCGCUU ((((((((((((....(((((((...((...))(((((((..(.................)..))))))).)))))))...)))))))).........((((..........)))))))) ( -35.63) >DroSec_CAF1 111763 120 - 1 AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCUCUUAAAAUCUACAAAUCUGCAAAGACUCGGCCAGUUUCUGCAGUCUGCAGUUGUUUGCCUAUUUAUAGUCAGGCGCUG ....((((((((....(((((((...((...))(((((((..(.................)..))))))).)))))))...))))))))((((.(((((.(((....))).))))))))) ( -36.63) >DroSim_CAF1 18022 120 - 1 AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCUCUUAAAAUCUACAAAUCUGCAAAGACUCGGCCAGUUUCUGCAGUCUGCAGUUGUUUGCCUAUUUAUAGUUAGGCGCUG .(((((((((((....(((((((...((...))(((((((..(.................)..))))))).)))))))...)))))))).........(((((........)))))))). ( -38.53) >DroEre_CAF1 34405 120 - 1 AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCGCUGAAAAUCUACAAAUCUGCAAAGACUCGGCCAGUUUCUGCAGUCUGCAGUUGUUUGCCUAUUUAUAGCUGGGCGCUU ..((((((((((....(((((((...((...))(((((((.((.((...........)).)).))))))).)))))))...))))))).(((((((..........)))))))))).... ( -42.20) >DroYak_CAF1 5213 119 - 1 AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCUCUGAAAAUCUACAAAUCUGCA-AGAGUCGGCCAGUUUCUGCAGUCUGCAGUUGUUUGCCUAUUUAUAGCUAGGCGCUU ((((((((((((....(((((((....(((((((((....))))))...))).....((....-.))....)))))))...)))))))).........(((((........))))))))) ( -38.30) >consensus AAGCCAGAUUGCUUUCAGCUGGCAGCCGGAUCGGAGUCUUCUCUUAAAAUCUACAAAUCUGCAAAGACUCGGCCAGUUUCUGCAGUCUGCAGUUGUUUGCCUAUUUAUAGUUAGGCGCUU .(((((((((((....(((((((...((...))(((((((..(.................)..))))))).)))))))...)))))))).........(((((........)))))))). (-34.67 = -35.31 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:53 2006