
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,056,072 – 21,056,347 |
| Length | 275 |
| Max. P | 0.925607 |

| Location | 21,056,072 – 21,056,192 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056072 120 - 23771897 AAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGGCCAGAUAUGCCAAUGGAGAUGA ....(((((((((((((......(((.....)))......))))))))((((..(((....(((((......)))))....)))..))))......(((.......))))))))...... ( -26.70) >DroSec_CAF1 111075 120 - 1 AAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGGCCAGAUAUGCCGAUGGAGAUGA ....(((((((((((((......(((.....)))......))))))))((((..(((....(((((......)))))....)))..))))......(((.......))))))))...... ( -27.10) >DroSim_CAF1 17343 120 - 1 AAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGACCAGAUAUGCCGAUGGAGAUGA ....(((((((((..((((((((..(((((((((.....)))))).)))...)))))).))(((((......))))).))......(((........)))......).))))))...... ( -25.80) >DroEre_CAF1 33634 120 - 1 AAGACCGUUGCAAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGGCCAGAUAUGCCGAUGGAGAUGA ..((...((((.....))))...))..............(((((((.(((((..(((....(((((......)))))....)))..))))).((..(((.......)))..))))))))) ( -26.90) >DroYak_CAF1 4532 120 - 1 AAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGGCCAGAUAUGCCGAUGCAGAUGA ......(((((.....(((((((...((((((((.....)))))).)))))))))......))))).....(((((((((...((......))...(((.......))))))))).))). ( -29.80) >consensus AAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUCAAAGUGGGUAAACAAAGGCCAGAUAUGCCGAUGGAGAUGA ....((((((((...((((((((..(((((((((.....)))))).)))...)))))).))(((((......))))).........(((........))).....)).))))))...... (-24.94 = -24.66 + -0.28)



| Location | 21,056,112 – 21,056,232 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -29.40 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056112 120 - 23771897 UGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCGAAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((..((....))...)))))))).))))..((((....((((((((......(((.....)))......)))))))).....))))....(((((......)))))... ( -34.90) >DroSec_CAF1 111115 120 - 1 UGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCGAAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((..((....))...)))))))).))))..((((....((((((((......(((.....)))......)))))))).....))))....(((((......)))))... ( -34.90) >DroSim_CAF1 17383 120 - 1 UGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCGAAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((..((....))...)))))))).))))..((((....((((((((......(((.....)))......)))))))).....))))....(((((......)))))... ( -34.90) >DroEre_CAF1 33674 120 - 1 UGGGGCGGAGAGGCAUCCAUUGACUUCUUCUGCUGCCGCGAAGACCGUUGCAAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((((.....)).)))).)))))).))((((......))))....((((((((..(((((((((.....)))))).)))...)))))).))(((((......)))))... ( -33.80) >DroYak_CAF1 4572 120 - 1 UGGGGAGGAGAGGCAUCCAUUGACUUCUUCUGCUGCCCCGAAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((((.....)).))))))))....))))..((((....((((((((......(((.....)))......)))))))).....))))....(((((......)))))... ( -33.90) >consensus UGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCGAAGACCGUUGCGAAAUGCAAAUAUCAACAAAUGAGCAAAUCGUUUCGUUAUUUGUUUGACAGCAACACAAACGUUGCAUC .((((((((((((..((....))...)))))))).))))................((((((((..(((((((((.....)))))).)))...)))))).))(((((......)))))... (-29.40 = -30.20 + 0.80)

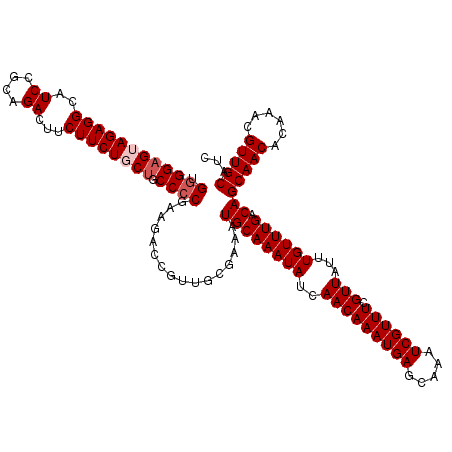

| Location | 21,056,192 – 21,056,312 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.54 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056192 120 - 23771897 UGGAGUUUAAGUCCCAAGAGGUCGUAGAAAUCGCAAAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUAUGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCG .(((((.((((((....((((((..(((..((((((......))))))..)))...).)).)))..)))))).)))))...((((((((((((..((....))...)))))))).)))). ( -38.20) >DroSec_CAF1 111195 120 - 1 UGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUAUGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCG .(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).)))))...((((((((((((..((....))...)))))))).)))). ( -42.90) >DroSim_CAF1 17463 120 - 1 UGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUAUGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCG .(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).)))))...((((((((((((..((....))...)))))))).)))). ( -42.90) >DroEre_CAF1 33754 120 - 1 UGGAGUUCAAGCCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCGCACUUGAGAUCUGCGUUCUUAUGGGGCGGAGAGGCAUCCAUUGACUUCUUCUGCUGCCGCG .(((((....((((((.(((..((((((..((((((......))))))..((((.((....)).))))))))))..))).))))))(((((((((.....)).))))))).))).))... ( -45.30) >DroYak_CAF1 4652 120 - 1 UGGAGUUCAAGCCCCAAGAGGUCGUAGAAAUCGCACAGUCCAUUGCGAAAUCUUUAGCACACUUGAGAUCUGCAUUCUUGUGGGGAGGAGAGGCAUCCAUUGACUUCUUCUGCUGCCCCG .(((((.....(((((.((((..(((((..(((((........)))))..((((.((....)).)))))))))..)))).)))))((((((((((.....)).))))))))))).))... ( -39.20) >consensus UGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUAUGGGGAGUAGAGGCAUCCGCAGACUUCUUCUGCUGCCCCG .(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).)))))...((((((((((((..((....))...)))))))).)))). (-33.26 = -34.54 + 1.28)


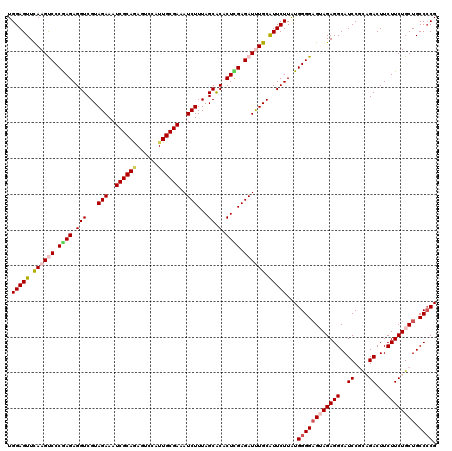
| Location | 21,056,232 – 21,056,347 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -31.22 |
| Energy contribution | -31.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21056232 115 - 23771897 GCGGGGAAUUUGCUCACAUA-----UGUGGUUUCCUCGUGUGGAGUUUAAGUCCCAAGAGGUCGUAGAAAUCGCAAAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUA (((((((....((((.((((-----((.((...)).)))))))))).....))))..((((((..(((..((((((......))))))..)))...).)).)))......)))....... ( -35.60) >DroSec_CAF1 111235 115 - 1 GCGGGGAAUAUGCUCACAUA-----UGUGGUUUCCUCGUGUGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUA ((((((((...((.(((...-----.)))))))))))))..(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).))))).. ( -38.70) >DroSim_CAF1 17503 115 - 1 GCGGGGAAUAUGCUCACAUA-----UGUGGUUUCCUCGUGUGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUA ((((((((...((.(((...-----.)))))))))))))..(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).))))).. ( -38.70) >DroEre_CAF1 33794 120 - 1 GCGGGGAAAAUGCUUACAUACUCGAUGUGGUUUCCUCGUGUGGAGUUCAAGCCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCGCACUUGAGAUCUGCGUUCUUA .(((((.....((((.(((((..((.......))...))))))))).....))))).(((..((((((..((((((......))))))..((((.((....)).))))))))))..))). ( -36.60) >DroYak_CAF1 4692 115 - 1 GCAGGGAAUAUGCUUACAUA-----UGUGAUUUCCUCGUGUGGAGUUCAAGCCCCAAGAGGUCGUAGAAAUCGCACAGUCCAUUGCGAAAUCUUUAGCACACUUGAGAUCUGCAUUCUUG ...(((.....((((.((((-----((.(.....).)))))))))).....)))((((((...(((((..(((((........)))))..((((.((....)).))))))))).)))))) ( -31.00) >consensus GCGGGGAAUAUGCUCACAUA_____UGUGGUUUCCUCGUGUGGAGUUCAAGUCCCGAGAGGUCGUAGAAAUCGCAGAGUCCAUUGCGAAAUCUUUAGCACACUCGAGAUUUGCAUUCUUA ((((((((.....(((((.......))))).))))))))..(((((.((((((.((((.(((...(((..((((((......))))))..)))...)).).)))).)))))).))))).. (-31.22 = -31.50 + 0.28)


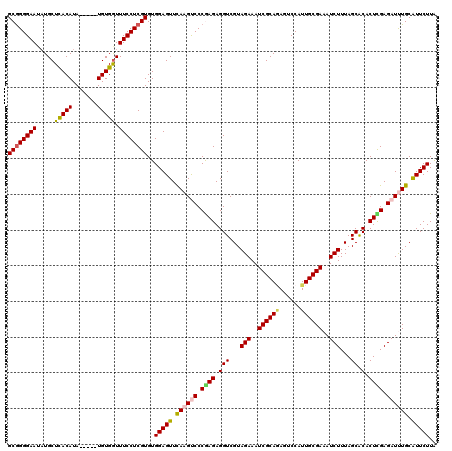
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:50 2006