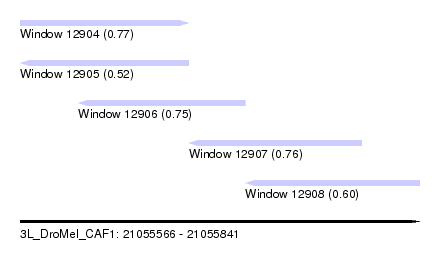
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,055,566 – 21,055,841 |
| Length | 275 |
| Max. P | 0.769395 |

| Location | 21,055,566 – 21,055,682 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -34.48 |
| Energy contribution | -35.68 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21055566 116 + 23771897 UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUGAGUUGGCCUCCUUGCUA----GUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGCCCCACGAAAGUCGCGGGGCGUUGCCAUU .........(((.((.....(((((((...((((((((....((....)----).....)))))))).((.(....).)))))))))...(((((.((.....)).))))))).)))... ( -40.00) >DroSec_CAF1 110585 116 + 1 UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUGAGUUGGCCUCCUUGCUG----GUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGCCCCACGAAAGUCGGGGGGCGUUGCCAUU .(((((((.(((((.......(((.......))).)))))(((((((((----(.....)))..(((((.....))))).)))))))....((((((....)).)))))))))))..... ( -43.60) >DroSim_CAF1 16843 116 + 1 UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUGAGUUGGCCUCCUUGCUG----GUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGCCCCACGAAAGUCGGGGGGCGUUGCCAUU .(((((((.(((((.......(((.......))).)))))(((((((((----(.....)))..(((((.....))))).)))))))....((((((....)).)))))))))))..... ( -43.60) >DroEre_CAF1 33135 119 + 1 UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUAAGCUGGCCUCCUUGCGAGUGAGUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGA-CCACGAAAGUCAGGGGGCGUUGCCAUU .(((((((.(((((...........((......)))))))((((((((.(((((((.....)))))))(((...)))..)))))))).....-((((....))..)).)))))))..... ( -36.70) >DroYak_CAF1 4051 115 + 1 UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUGAGUU-GCCUCCUUGCGC----GUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGCCCCACGAAAGUCGGGGGGCGUUGCCAUU .(((((((.(((.........(((.......)))..-)))((((((((.----(((.....)))(((((.....)))))))))))))....((((((....)).)))))))))))..... ( -38.20) >consensus UCAAUGCUGGGCCAUAAAUAUCUCUGCACUUGAGUUGGCCUCCUUGCUA____GUUUUUCCGACUCAUGUGAGCCAUGACGCAGGGAAAAGCCCCACGAAAGUCGGGGGGCGUUGCCAUU .(((((((.(((((.......(((.......))).)))))(((((((......(((.....)))(((((.....))))).)))))))....((((((....)).)))))))))))..... (-34.48 = -35.68 + 1.20)


| Location | 21,055,566 – 21,055,682 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.58 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21055566 116 - 23771897 AAUGGCAACGCCCCGCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----UAGCAAGGAGGCCAACUCAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ...(((...((((((((.....))))))))((((((..((.(((((.....))))))).)))))).(----(.(((..(((.....)))...)))))...........)))......... ( -40.90) >DroSec_CAF1 110585 116 - 1 AAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----CAGCAAGGAGGCCAACUCAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ..((((...(((((.((.....)).)))))..((((.(((.....(((((....)))))((.....)----).))).)))))))).....(((((..................))))).. ( -37.77) >DroSim_CAF1 16843 116 - 1 AAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----CAGCAAGGAGGCCAACUCAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ..((((...(((((.((.....)).)))))..((((.(((.....(((((....)))))((.....)----).))).)))))))).....(((((..................))))).. ( -37.77) >DroEre_CAF1 33135 119 - 1 AAUGGCAACGCCCCCUGACUUUCGUGG-UCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAACUCACUCGCAAGGAGGCCAGCUUAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ...(((...(((.(((.......((((-..((((((..((.(((((.....))))))).))))))..)))).....))).)))..((......)).............)))......... ( -30.30) >DroYak_CAF1 4051 115 - 1 AAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----GCGCAAGGAGGC-AACUCAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ...(((...(((((.((.....)).))))).((((...((((..((((((....)))))).....))----))(((..(((..-..)))...))).))))........)))......... ( -41.10) >consensus AAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC____CAGCAAGGAGGCCAACUCAAGUGCAGAGAUAUUUAUGGCCCAGCAUUGA ...(((...(((((.((.....)).)))))((((((..((.(((((.....))))))).))))))........(((..(((.....)))...))).............)))......... (-33.50 = -33.58 + 0.08)

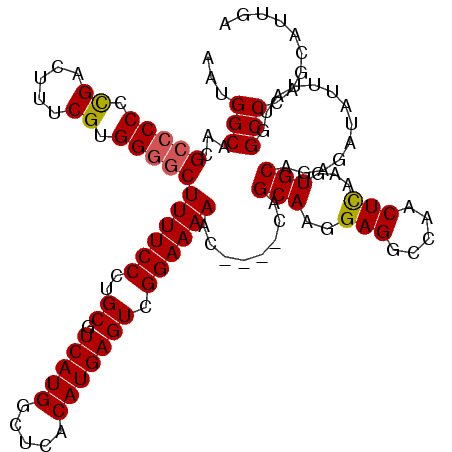
| Location | 21,055,606 – 21,055,721 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -30.76 |
| Energy contribution | -31.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21055606 115 - 23771897 CCCAUCGUCCACACCAG-ACACGCCCCCUCGGCGCGAAUCAAUGGCAACGCCCCGCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----UAGCAAGGA .((.(((((.......)-)).((((.....)))).)).......((...((((((((.....))))))))((((((..((.(((((.....))))))).))))))..----..))..)). ( -40.50) >DroSec_CAF1 110625 114 - 1 CCCAUCGUCCACACCAG-ACACGCC-CCUCGGCGCGAAUCAAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----CAGCAAGGA .((((.(((.......)-)).((((-....)))).......))))....(((((.((.....)).)))))...(((.(((.....(((((....)))))((.....)----).))).))) ( -37.00) >DroSim_CAF1 16883 114 - 1 CCCAUCGUCCACACCAG-GCACGCC-CCUCGGCGCGAAUCAAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----CAGCAAGGA .............((..-((.((((-....))))........(((....(((((.((.....)).)))))((((((..((.(((((.....))))))).)))))).)----))))..)). ( -37.20) >DroEre_CAF1 33175 119 - 1 CCCAUCGUCCACACCAGGCCACGCCCCCCAUGCGCGAAUCAAUGGCAACGCCCCCUGACUUUCGUGG-UCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAACUCACUCGCAAGGA ...............(((((((((.......))..((((((..(((...)))...)))..)))))))-)))..(((.((((...((((((....))))))((.....))...)))).))) ( -29.30) >DroYak_CAF1 4090 116 - 1 CCCAUCGUCCACACCAAGACACGCCCCCUAUGCGCGAAUCAAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC----GCGCAAGGA ......(((........)))......(((.((((((.......(....)(((((.((.....)).)))))((((((..((.(((((.....))))))).)))))).)----)))))))). ( -41.50) >consensus CCCAUCGUCCACACCAG_ACACGCCCCCUCGGCGCGAAUCAAUGGCAACGCCCCCCGACUUUCGUGGGGCUUUUCCCUGCGUCAUGGCUCACAUGAGUCGGAAAAAC____CAGCAAGGA .......(((.........(.(((.......))).)........((...(((((.((.....)).)))))((((((..((.(((((.....))))))).))))))........))..))) (-30.76 = -31.00 + 0.24)

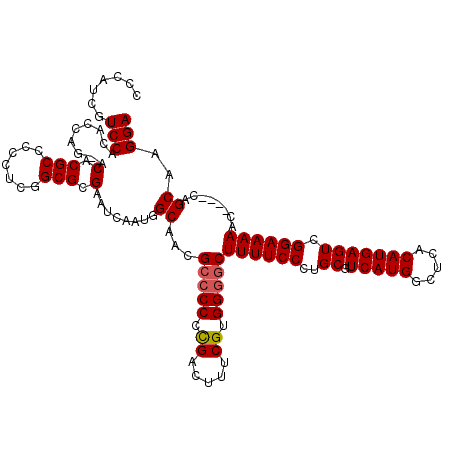
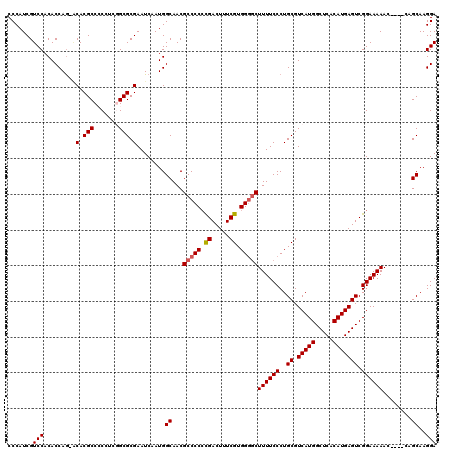
| Location | 21,055,682 – 21,055,801 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.79 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21055682 119 - 23771897 UUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUUGUAUUAUGAGCGCCGUGGAUUGCUCAUACGCCCCAUCGUCCACACCAG-ACACGCCCCCUCGGCGCGAAUC ..........((((.((((((((((....))))(((......)))))))))...))))((((((.((...((......))......(((.......)-))......)).))))))..... ( -31.80) >DroSec_CAF1 110701 118 - 1 UUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUUGUAUUAUGAGCGCCGUGGAUUACUCAUACGCCCCAUCGUCCACACCAG-ACACGCC-CCUCGGCGCGAAUC ..........((((.((((((((((....))))(((......)))))))))...))))(((((((((((.................))))))....(-(......-..)))))))..... ( -29.83) >DroSim_CAF1 16959 118 - 1 UUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUGGUAUUAUGAGCGCCGUGGAUUACUCAUACGCCCCAUCGUCCACACCAG-GCACGCC-CCUCGGCGCGAAUC .............((.(..............(((((...((.((((((.((...)).))))))((((((.................)))))).)).)-))))(((-....)))).))... ( -31.43) >DroEre_CAF1 33254 119 - 1 UUAAUUAAACUCAUCACAAACACUU-UGCGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGCCCCAUCGUCCACACCAGGCCACGCCCCCCAUGCGCGAAUC .......................((-((((.(((.....((.(((((((((...)))))))))((((((.................)))))).)).(((...)))...))).)))))).. ( -31.83) >DroYak_CAF1 4166 120 - 1 UUAAUUAAACGCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGCCCCAUCGUCCACACCAAGACACGCCCCCUAUGCGCGAAUC .........(((........(((((....))))).((.(((.(((((((((...)))))))))((((((.................))))))..............))).)).))).... ( -30.73) >consensus UUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGCCCCAUCGUCCACACCAG_ACACGCCCCCUCGGCGCGAAUC ....................(((((....))))).....((.(((((((((...)))))))))((((((.................)))))).))....(.(((.......))).).... (-26.75 = -26.79 + 0.04)

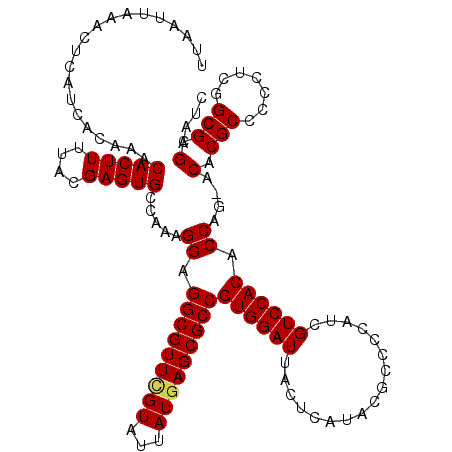
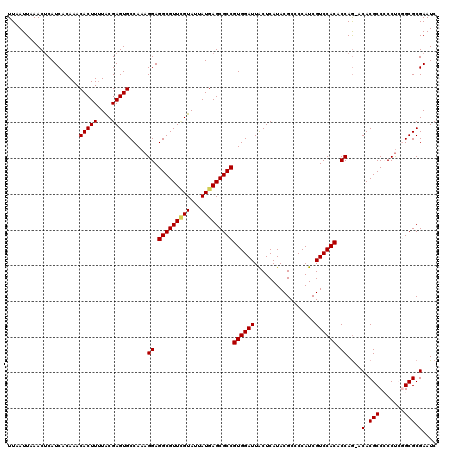
| Location | 21,055,721 – 21,055,841 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -28.88 |
| Energy contribution | -28.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21055721 120 - 23771897 UCUGCUGGGCGAAAUGCAAAUGCAAAUCCCUCGCAGAUCUUUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUUGUAUUAUGAGCGCCGUGGAUUGCUCAUACGC (((((.(((.((..(((....)))..))))).)))))..................((((((((((....))))(((......)))))))))..((((((((........))))))))... ( -35.10) >DroSec_CAF1 110739 114 - 1 UCUGCUGGGCGAAAUG------CAAAUCCCUCGCAGAUCUUUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUUGUAUUAUGAGCGCCGUGGAUUACUCAUACGC (((((.(((.((....------....))))).)))))................................((((((((.....((((((..(...)..)))))).)))..)))))...... ( -28.80) >DroSim_CAF1 16997 114 - 1 UCUGCUGGGCGAAAUG------CAAAUCCCUCGCAGAUCUUUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUGGUAUUAUGAGCGCCGUGGAUUACUCAUACGC (((((.(((.((....------....))))).)))))................................((((((((.....((((((.((...)).)))))).)))..)))))...... ( -28.10) >DroEre_CAF1 33294 113 - 1 UCUGCCUGGCGGAAUG------CAAAUCCCUCGCAGAUCUUUAAUUAAACUCAUCACAAACACUU-UGCGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGC ((..(.(((((((...------....)))((((((((..........................))-)))))).)))).....(((((((((...))))))))))..))............ ( -31.97) >DroYak_CAF1 4206 114 - 1 UCUGCUGGGCGGAAUG------CAAAUCCCUCGCAGGUCUUUAAUUAAACGCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGC (((((.(((.(((...------....)))))))))))................................((((((((.....(((((((((...))))))))).)))..)))))...... ( -31.50) >consensus UCUGCUGGGCGAAAUG______CAAAUCCCUCGCAGAUCUUUAAUUAAACUCAUCACAAACACUUUUACGAGUGCCAAAGGAGGCGUUCGUAUUAUGAGCGCCGUGGAUUACUCAUACGC (((((.(((.((..(((....)))..))))).)))))................................((((.(((.....(((((((((...))))))))).)))...))))...... (-28.88 = -28.76 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:46 2006