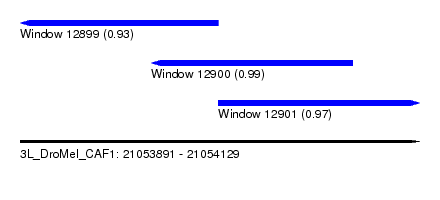
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,053,891 – 21,054,129 |
| Length | 238 |
| Max. P | 0.987361 |

| Location | 21,053,891 – 21,054,009 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.94 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21053891 118 - 23771897 GGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA-AAUUGUGGGGUACUUUUCUGCAAUGAUUCGCCAGUAUUUUUAAAUACUAA-AAAUACAAUUUGCAUACUCGUAUUACCU .((.(((((...((((((((((.((......)).))))).-.((((..(((.....)))..))))...))))).(((((((((.....)))-)))))))))))))............... ( -32.50) >DroSec_CAF1 108921 119 - 1 GGACAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCAAAAUUGUGGGGUACUUUUCUGCGAUGAUUCGCCAGUAUUUUUAAAUACUAA-AAAUAUAAUUUGCAUACUCGUAUUACCU .(.((((((...((((((((((.((......)).)))))...((((..(((.....)))..))))...))))).(((((((((.....)))-)))))))))))))............... ( -29.90) >DroSim_CAF1 15185 118 - 1 GGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA-AAUUGUGGGGUACUUUUCUGCAAUGAUUCGCCAGUAUUUUUAAAUACUAA-AAUCACAAUUUGCAUACUCGUAUUACCU ((((..((.....))..))))((((((.....))))))((-(((((((((((.....((......))...)))((((((....))))))..-..))))))))))................ ( -31.40) >DroEre_CAF1 31466 113 - 1 GGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA-AAUUGUGGGGUACUUUUCUGCAAUGAUUCGCGAGUAUUUUUAAAUAGUAAAAAAUACAAUUUGCAU------AUUACCU ((((((.(((..((((....))))...)))...)))))).-.((((..(((.....)))..)))).....((((((((((((........))))))))...))))..------....... ( -30.90) >DroYak_CAF1 2376 113 - 1 GGCAAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA-AAUUGUGGGGUACUUUUCUGCAAUGAUUCGCCAGUAUUUUUAAAUAGUAGAAAAUACAAUUUGCAU------AUUACCU .(((((......((((((((((.((......)).))))).-.((((..(((.....)))..))))...))))).((((((((........))))))))..)))))..------....... ( -34.10) >consensus GGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA_AAUUGUGGGGUACUUUUCUGCAAUGAUUCGCCAGUAUUUUUAAAUACUAA_AAAUACAAUUUGCAUACUCGUAUUACCU ...........(((((((((.((((((.....))))))....((((..(((.....)))..)))).....))).(((((((((.....))).))))))................)))))) (-25.82 = -25.94 + 0.12)

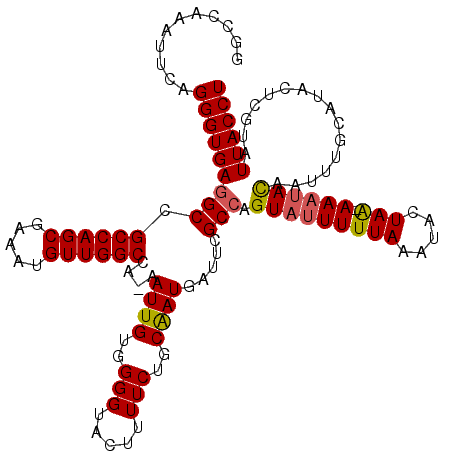

| Location | 21,053,969 – 21,054,089 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -37.46 |
| Consensus MFE | -33.40 |
| Energy contribution | -34.08 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21053969 120 - 23771897 AAACUGCUGGCCCAGGCGAAAAAUCUGCCAAACAAUUGUUAUCUGCCAGCGAACAGAGACAAUUGCCGCCUUUGUCUUUUGGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA ....(((((((...((((.......)))).(((....)))....)))))))...((((((((.........))))))))(((((((.(((..((((....))))...)))...))))))) ( -39.90) >DroSec_CAF1 109000 120 - 1 AAACUGCUGGCCUAGGCGAAAAAUCUGCCAAACAAUUGUUAUCUGCCAGCGAACAGAGACAAUUGCCACCUUUGUCUUUUGGACAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA ........(((((.((((.......))))...((((((((.((((........)))))))))))).((((((((((.....))))).....))))))))))((((((.....)))))).. ( -42.80) >DroSim_CAF1 15263 115 - 1 A-----UUGGCCCAGGCGAAAAAUCUGCCAAACAAUUGUUAUCUGCCAGCGAACAGAGACAAUUGCCGCCUUUGUCUUUUGGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA .-----..((((..((((.......))))...((((((((.((((........)))))))))))).((((((.(((....))).......)))))).))))((((((.....)))))).. ( -36.90) >DroEre_CAF1 31539 119 - 1 AAACUGC-GGCCCAGGCUAAAAAUCUGCCAAACAAUUGUUAUCUACCAGCGCACAGAGACAAUUGCCGCCUUUGUCUUUUGGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA .....((-(((...(((.........)))....(((((((.(((..........)))))))))))))))..........(((((((.(((..((((....))))...)))...))))))) ( -34.80) >DroYak_CAF1 2449 120 - 1 AAACUGCUGGCCCAGGCUAAAAAUCUACCAAACAAUUGUUAUCUACCAGCGAACGGAGACAAUUGCCGCCUUUGUCUUUUGGCAAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA ....(((((((...((((..............((((((((.(((..........))))))))))).((((((((((....)))))......))))).)))))))))))............ ( -32.90) >consensus AAACUGCUGGCCCAGGCGAAAAAUCUGCCAAACAAUUGUUAUCUGCCAGCGAACAGAGACAAUUGCCGCCUUUGUCUUUUGGCCAAAUUCAGGGUGAGGCCGCCAGCGAAAUGUUGGCCA ........((((..((((.......))))...((((((((.((((........)))))))))))).((((((.(...(((....)))..))))))).))))((((((.....)))))).. (-33.40 = -34.08 + 0.68)


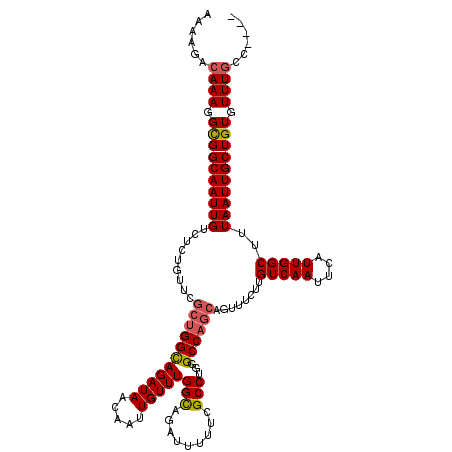
| Location | 21,054,009 – 21,054,129 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.37 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21054009 120 + 23771897 AAAAGACAAAGGCGGCAAUUGUCUCUGUUCGCUGGCAGAUAACAAUUGUUUGGCAGAUUUUUCGCCUGGGCCAGCAGUUUCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUUGCCGUUU ..........((((((((((((.(((((......)))))..))))))))).(((.((....))))).((((((((((((....(((((....)))))...)))))))).))))))).... ( -37.90) >DroSec_CAF1 109040 116 + 1 AAAAGACAAAGGUGGCAAUUGUCUCUGUUCGCUGGCAGAUAACAAUUGUUUGGCAGAUUUUUCGCCUAGGCCAGCAGUUUCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUUGCC---- .............(((((((((.(((((......)))))..))))))))).(((((((.....((....))((((((((....(((((....)))))...)))))))).)))))))---- ( -34.70) >DroSim_CAF1 15303 111 + 1 AAAAGACAAAGGCGGCAAUUGUCUCUGUUCGCUGGCAGAUAACAAUUGUUUGGCAGAUUUUUCGCCUGGGCCAA-----UCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUUGCC---- ......((((.((((((((((..(((((......)))))...((((...((((((((((....((....)).))-----)).))))))...))))....)))))))))).))))..---- ( -32.50) >DroEre_CAF1 31579 116 + 1 AAAAGACAAAGGCGGCAAUUGUCUCUGUGCGCUGGUAGAUAACAAUUGUUUGGCAGAUUUUUAGCCUGGGCC-GCAGUUUCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUUGCCG--- ....(.((((.((((((((((...(((((..(((.((((((.....)))))).))).......((....)))-))))......(((((....)))))..)))))))))).)))).).--- ( -32.90) >DroYak_CAF1 2489 113 + 1 AAAAGACAAAGGCGGCAAUUGUCUCCGUUCGCUGGUAGAUAACAAUUGUUUGGUAGAUUUUUAGCCUGGGCCAGCAGUUUCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUU------- ...(((((..((((((((..(((((((..((...((.....))...))..))).))))..)).)))...)))(((((((....(((((....)))))...))))))).)))))------- ( -29.20) >consensus AAAAGACAAAGGCGGCAAUUGUCUCUGUUCGCUGGCAGAUAACAAUUGUUUGGCAGAUUUUUCGCCUGGGCCAGCAGUUUCUUGUCAAUUCAUUGGCUUUAAUUGCUGUGUUUGCC____ ......((((.((((((((((.........(((((((((((.....)))))(((.........)))...))))))........(((((....)))))..)))))))))).))))...... (-29.40 = -29.64 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:39 2006