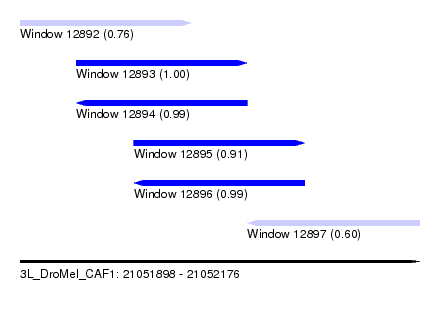
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,051,898 – 21,052,176 |
| Length | 278 |
| Max. P | 0.998709 |

| Location | 21,051,898 – 21,052,017 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.15 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051898 119 + 23771897 AGUUUUUGGUCU-UUGAAAACGUCAUCGUGUCGCAGAUAAAAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAAU .......(((((-(((.....((((((........)))...................((...))..............)))......))))))))...(((((((....))))))).... ( -24.50) >DroSec_CAF1 107048 119 + 1 AGUUUUUGGUCUUUUGAAAACGUCAUCGUGUCGCAGAUAAAAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA- .(((((..(....)..)))))(.(((..((((.........................((...))......(.....).))))..))))..........(((((((....)))))))...- ( -21.80) >DroSim_CAF1 13261 119 + 1 AGUUUUUGGUCUUUUGAAAACGUCAUCGUGUCGCAGAUAAAAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAAACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA- .((((((..((((((......(((...((((.....................)))).)))......))))))..)).)))).................(((((((....)))))))...- ( -20.90) >DroEre_CAF1 29470 118 + 1 AGUUUUUGGUCU-UUGAAAACGUCAUCGUGUCGCAGAUAAAAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA- .......(((((-(((.....((((((........)))...................((...))..............)))......))))))))...(((((((....)))))))...- ( -24.50) >DroYak_CAF1 526 117 + 1 AGUUUUUGGUCU-UUGAAAACGUCAUCGUGUCGCAGACAAAAUAACAAACAAACACCGGCCCCCAA-AAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA- .......(((((-(((.....(((....((((...))))..................((...))..-...........)))......))))))))...(((((((....)))))))...- ( -25.40) >consensus AGUUUUUGGUCU_UUGAAAACGUCAUCGUGUCGCAGAUAAAAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA_ .(((((..(....)..)))))(.(((..((((.........................((...))......(.....).))))..))))..........(((((((....))))))).... (-19.98 = -20.18 + 0.20)



| Location | 21,051,937 – 21,052,056 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.61 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051937 119 + 23771897 AAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAAUAUCUCAUCUCCUGCGUUUUCCCAGGCUCAAAGGAAAAA-A ...............((((((..(......).......(..((((((((.((((....(((((((....))))))).........))))..))))))))..).))))....)).....-. ( -20.92) >DroSec_CAF1 107088 117 + 1 AAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCUCAGGCUCAAAGGAAAAA-A ...............((((((..(......).......((.((((((((.((((....(((((((....)))))))...--....))))..)))))))).)).))))....)).....-. ( -25.70) >DroSim_CAF1 13301 117 + 1 AAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAAACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAA-A ...............((((((..(......)..........((((((((.((((....(((((((....)))))))...--....))))..))))))))....))))....)).....-. ( -23.00) >DroEre_CAF1 29509 117 + 1 AAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUU-UCUCAGGCUCAAAGGAAACGAG .................((((..(......).......((..(((((((.((((....(((((((....)))))))...--....))))..))))))-).)).))))....(....)... ( -23.60) >DroYak_CAF1 565 117 + 1 AAUAACAAACAAACACCGGCCCCCAA-AAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAGAG ...............((((((.....-...(.....).(..((((((((.((((....(((((((....)))))))...--....))))..))))))))..).))))....))....... ( -23.10) >consensus AAUAACAAACAAACACCGGCCCCCAAAAAAGAAAAACAGACAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA__UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAA_A ...............((((((..(......)..........((((((((.((((....(((((((....))))))).........))))..))))))))....))))....))....... (-21.04 = -21.24 + 0.20)



| Location | 21,051,937 – 21,052,056 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.61 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.40 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051937 119 - 23771897 U-UUUUUCCUUUGAGCCUGGGAAAACGCAGGAGAUGAGAUAUUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUCUGUUUUUCUUUUUUGGGGGCCGGUGUUUGUUUGUUAUU .-.....((((..((...(((((((((.(.((((((((((.(((((((((......))))))))).))))...)))))).).)))))))))..))..))))................... ( -34.60) >DroSec_CAF1 107088 117 - 1 U-UUUUUCCUUUGAGCCUGAGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUCUGUUUUUCUUUUUUGGGGGCCGGUGUUUGUUUGUUAUU .-.....((((..((...(((((((.((((((((.((((--(((((((((......)))))))..)))))).....)))).))))))))))).))..))))................... ( -31.90) >DroSim_CAF1 13301 117 - 1 U-UUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUUUGUUUUUCUUUUUUGGGGGCCGGUGUUUGUUUGUUAUU .-.....((((..((...((((((((((((((((.((..--(((((((((......)))))))))..)))))))....)))..))))))))..))..))))................... ( -32.90) >DroEre_CAF1 29509 117 - 1 CUCGUUUCCUUUGAGCCUGAGA-AACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUCUGUUUUUCUUUUUUGGGGGCCGGUGUUUGUUUGUUAUU .(((...((((..((...((((-((.((((((((.((((--(((((((((......)))))))..)))))).....)))).)))).)))))).))..)))).)))............... ( -33.40) >DroYak_CAF1 565 117 - 1 CUCUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUCUGUUUUUCUUU-UUGGGGGCCGGUGUUUGUUUGUUAUU .......((((..((...((((((((((((((((.((..--(((((((((......)))))))))..)))))))....)))..)))))))).)-)..))))................... ( -32.50) >consensus U_UUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA__UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUGUCUGUUUUUCUUUUUUGGGGGCCGGUGUUUGUUUGUUAUU .......((((..((...(((((((.((((((((.((((..(((((((((......)))))))))..)))).....)))).))))))))))).))..))))................... (-29.20 = -29.40 + 0.20)



| Location | 21,051,977 – 21,052,096 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051977 119 + 23771897 CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAAUAUCUCAUCUCCUGCGUUUUCCCAGGCUCAAAGGAAAAA-ACUUUUCUUCUCACUCGCUUCAAAGCUGAAUAAAAACACUU .((((((((.((((....(((((((....))))))).........))))..)))))))).......((((((((((..-..))))))).......(((....))))))............ ( -22.02) >DroSec_CAF1 107128 117 + 1 CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCUCAGGCUCAAAGGAAAAA-ACUUUUCUUCUCACUCGCUUCAAAGCUGAAUAAAUACACUU .((((((((.((((....(((((((....)))))))...--....))))..)))))))).......((((((((((..-..))))))).......(((....))))))............ ( -24.30) >DroSim_CAF1 13341 117 + 1 CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAA-ACUUUUCUUCUCACUCGCUUCAAAGCUGAAUAAAUACACUU .((((((((.((((....(((((((....)))))))...--....))))..)))))))).......((((((((((..-..))))))).......(((....))))))............ ( -24.30) >DroEre_CAF1 29549 117 + 1 CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUU-UCUCAGGCUCAAAGGAAACGAGAUUUUCUUGUCACUCGCUUCAAAGCAGAAUAAAUACACUU ..................(((((((....)))))))...--.....((((((.((((-(((..........))))))).(((......)))...........))))))............ ( -27.50) >DroYak_CAF1 604 118 + 1 CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA--UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAGAGGUUUUCUUGUCACUCGCUUCAAAGCAGAAUAAAUACACUU ..................(((((((....)))))))...--.....((((((...((((((..........))))))(((((.............)))))..))))))............ ( -26.72) >consensus CAAAAUGCAAAGAUCUCCGGCUGAUUGAUGUCAGCCAAA__UCUCAUCUGCUGCGUUUUCCCAGGCUCAAAGGAAAAA_ACUUUUCUUCUCACUCGCUUCAAAGCUGAAUAAAUACACUU .((((((((.((((....(((((((....))))))).........))))..))))))))..........(((((((.....))))))).......(((....)))............... (-21.96 = -22.16 + 0.20)



| Location | 21,051,977 – 21,052,096 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -29.22 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051977 119 - 23771897 AAGUGUUUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAGU-UUUUUCCUUUGAGCCUGGGAAAACGCAGGAGAUGAGAUAUUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ...(.(((((((((.(((....)))))))))))).)...((-.(((((((........))))))).))..((((((((((.(((((((((......))))))))).))))...)))))). ( -36.00) >DroSec_CAF1 107128 117 - 1 AAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAGU-UUUUUCCUUUGAGCCUGAGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ..((((.(((.(((((.((..(((.....(((((((....)-)))))))))..)).)))))))))))).(((((.((..--(((((((((......)))))))))..)))))))...... ( -35.70) >DroSim_CAF1 13341 117 - 1 AAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAGU-UUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ..((((.(((((((.(((....)))))))))).........-..((((((........)))))))))).(((((.((..--(((((((((......)))))))))..)))))))...... ( -36.80) >DroEre_CAF1 29549 117 - 1 AAGUGUAUUUAUUCUGCUUUGAAGCGAGUGACAAGAAAAUCUCGUUUCCUUUGAGCCUGAGA-AACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ..((((.(((.....((((.((((((((............))))))))....))))...)))-.)))).(((((.((..--(((((((((......)))))))))..)))))))...... ( -37.80) >DroYak_CAF1 604 118 - 1 AAGUGUAUUUAUUCUGCUUUGAAGCGAGUGACAAGAAAACCUCUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA--UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ..((((..((((((.(((....))))))))).............((((((........)))))))))).(((((.((..--(((((((((......)))))))))..)))))))...... ( -35.80) >consensus AAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAGU_UUUUUCCUUUGAGCCUGGGAAAACGCAGCAGAUGAGA__UUUGGCUGACAUCAAUCAGCCGGAGAUCUUUGCAUUUUG ..((((..((((((.(((....))))))))).............((((((........)))))))))).(((((.((....(((((((((......)))))))))..)))))))...... (-29.22 = -30.02 + 0.80)



| Location | 21,052,056 – 21,052,176 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.63 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21052056 120 - 23771897 UAAAACACGAAACUUUUGCAUUUAAUUGGGCAAAUUUGCGCGAGUCAAGCAAGGGAUAUGAGCAAAGUUGAGGUGCAACAAAGUGUUUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAG .....(((....(((((((......(((.(((....))).))).....)))))))...((..((((((((((....(((.....)))....))))))))))...)).))).......... ( -29.10) >DroSec_CAF1 107205 119 - 1 UAAAACACGAAAAU-UUGCAUUUAAUUGGGCAAGUUUGCGCGAGCCAAGCAAGGGAUAUGAGCAAAGUUAAGGAGCAAAAAAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAG ....((((.....(-((((.(((((((..((..((((((....).))))).(......)..))..)))))))..)))))...)))).(((((((.(((....))))))))))........ ( -25.60) >DroSim_CAF1 13418 119 - 1 UAAAACACGAAAAU-UUGCAUUUAAUUGGGCAAGUUUGCGCGAGUCGAGCAAGGGAUAUGAGCAAAGUUGAGGUGCAAAAAAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAG ....((((.....(-((((((((.......(((.(((((.((.(((........))).)).))))).))))))))))))...)))).(((((((.(((....))))))))))........ ( -27.91) >DroEre_CAF1 29626 118 - 1 UAAAACAUGAAA-U-UUGCAUUUAAUUGAGCAAGUUUGCCCGAGUCAAGCGGGGGAUACGAGAAAAGUUGAGGUGCAACAAAGUGUAUUUAUUCUGCUUUGAAGCGAGUGACAAGAAAAU ....((((....-.-((((((((((((...(..((((.((((.......)))).)).))..)...)))))).))))))....))))..((((((.(((....)))))))))......... ( -25.80) >DroYak_CAF1 682 118 - 1 UAAAACAAGAAA-U-UUGCAUUUAAUUGGGCAAGUUUGCGCGAGUCAAGCAAGGGAUACGAGAAAAGUUGCGAUGCGACAAAGUGUAUUUAUUCUGCUUUGAAGCGAGUGACAAGAAAAC ........(..(-(-((((.(((((...((((.((((((....).)))))...(((((((......(((((...)))))....)))))))....)))))))))))))))..)........ ( -25.10) >consensus UAAAACACGAAA_U_UUGCAUUUAAUUGGGCAAGUUUGCGCGAGUCAAGCAAGGGAUAUGAGCAAAGUUGAGGUGCAACAAAGUGUAUUUAUUCAGCUUUGAAGCGAGUGAGAAGAAAAG ....((((.......((((((((.......(((.(((((.((.(((........))).)).))))).)))))))))))....))))..((((((.(((....)))))))))......... (-19.91 = -20.63 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:35 2006