
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,051,617 – 21,051,797 |
| Length | 180 |
| Max. P | 0.935649 |

| Location | 21,051,617 – 21,051,723 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.78 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051617 106 - 23771897 AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA-AACAACGAUAGA-------------CGAAAGACGAGACGCGUCUGUCAAUGACAC ......(((........))).((((.........((((.........)))).............-......((((((-------------((...(......).))))))))..)))).. ( -23.10) >DroSec_CAF1 106768 105 - 1 AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAA-AGCCGAUUCCGAAUCCAA-AGCUACGAUAGA-------------CGAAAGACGAGACGCGUCUGUCAAUGACAC ......(((........))).((((....(((..((((.......-.)))).....(....)..-.)))..((((((-------------((...(......).))))))))..)))).. ( -24.20) >DroSim_CAF1 12980 106 - 1 AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA-AGCUACGAUAGA-------------CGAAAGACGAGACGCGUCUGUCAAUGACAC ......(((........))).((((....(((..((((.........)))).....(....)..-.)))..((((((-------------((...(......).))))))))..)))).. ( -23.40) >DroEre_CAF1 29165 107 - 1 AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGACUCCGAAACCAAAAACUGCGACAGA-------------CGAAAGACGAGACGCGUCUGUCAAUGACAC ......(((........))).((((.........((((.........))))....................((((((-------------((...(......).))))))))..)))).. ( -25.00) >DroYak_CAF1 227 120 - 1 AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAACCAAAAACUGCGAUAGACCAAGACGAAAUACGAAAGACGAGACGCGUCUGUCAAUGACAC ................((((((((..........((((.........))))................(((...))).....))).......((.(((((.....))))).))..))))). ( -21.30) >consensus AAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA_AACUACGAUAGA_____________CGAAAGACGAGACGCGUCUGUCAAUGACAC ......(((........))).((((.........((((.........))))........................................((.(((((.....))))).))..)))).. (-19.50 = -19.50 + 0.00)

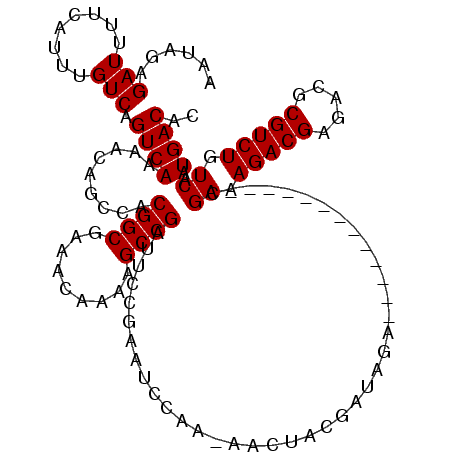
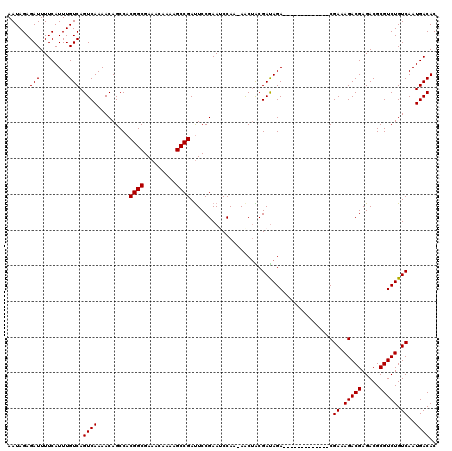
| Location | 21,051,647 – 21,051,757 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.95 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.76 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051647 110 - 23771897 ------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA-AACAACGAUAGA--- ------((((.....((((((((((((.(((((....))))...(((...)))....).))))))))..)))).((((.........)))).............-......))))..--- ( -27.70) >DroSec_CAF1 106798 109 - 1 ------UGUCUCUUCGUCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAA-AGCCGAUUCCGAAUCCAA-AGCUACGAUAGA--- ------..(((..((((.(((((((((.(((((....))))...(((...)))....).))))))))).(((..((((.......-.)))).....(....)..-.))))))).)))--- ( -24.40) >DroSim_CAF1 13010 110 - 1 ------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA-AGCUACGAUAGA--- ------((((.....((((((((((((.(((((....))))...(((...)))....).))))))))..)))).((((.........)))).............-......))))..--- ( -27.70) >DroEre_CAF1 29195 117 - 1 UGUGUUUGUUUCUUUGGCCUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGACUCCGAAACCAAAAACUGCGACAGA--- .(((((((((((..((((.((((((((.(((((....))))...(((...)))....).))))))))...))))((((.........)))).....)))))...)))).))......--- ( -29.70) >DroYak_CAF1 267 114 - 1 ------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAACCAAAAACUGCGAUAGACCA ------.((((..((((((((((((((.(((((....))))...(((...)))....).))))))))..)))).((((.........))))....................)).)))).. ( -30.20) >consensus ______UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGAAACAAAAGCCGAUUCCGAAUCCAA_AACUACGAUAGA___ ......((((.....((((((((((((.(((((....))))...(((...)))....).))))))))..)))).((((.........))))....................))))..... (-25.68 = -25.76 + 0.08)

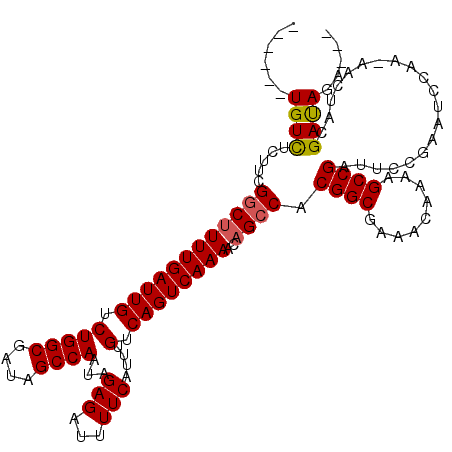
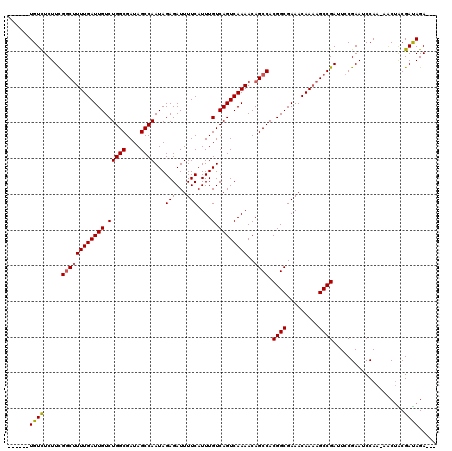
| Location | 21,051,683 – 21,051,797 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -29.35 |
| Energy contribution | -29.63 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051683 114 + 23771897 UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAAGCCGAAGAGACA------AACAGCCAUGAGAAGUCGUCUGUCGACGGUUGCAAACUUU ..(((((((((((((((.....))........(((((.((((((.................))))))))))).)------))))))))))...(..((((....))))..)))....... ( -32.93) >DroSec_CAF1 106833 114 + 1 UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAAGACGAAGAGACA------CACAGCCAUGAGAAGUCGUCUGUCGCCGGUUGCAAACCUU ..(((((((((((((((.....))))......(((((.(((((....)))))....((....))...)))))..------.)))))))))(((.....)))...)).(((.....))).. ( -26.50) >DroSim_CAF1 13046 114 + 1 UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAAGCCGAAGAGACA------AACAGCCAUGAGAAGUCGUCUGUCGACGGUUGCAAACUUU ..(((((((((((((((.....))........(((((.((((((.................))))))))))).)------))))))))))...(..((((....))))..)))....... ( -32.93) >DroEre_CAF1 29232 120 + 1 UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAGGCCAAAGAAACAAACACAAACAGCCAUGAGAAGUCGUCUGUCGACGGUUGCAAACUUU ..(((((((((((((((..((....((....)).(((.((((((.................)))))))))..))..)).)))))))))))...(..((((....))))..)))....... ( -30.03) >DroYak_CAF1 307 114 + 1 UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAAGCCGAAGAGACA------AACAGCCAUGAGAAGUCGUCUGUCGACGGUUGCAAACUUU ..(((((((((((((((.....))........(((((.((((((.................))))))))))).)------))))))))))...(..((((....))))..)))....... ( -32.93) >consensus UCGCCGUGGCUGUUUUGACUGACAAAUGAAAAUCUCUAUUGGCUAUCGCCAGACAAUCAAAAGCCGAAGAGACA______AACAGCCAUGAGAAGUCGUCUGUCGACGGUUGCAAACUUU ..(((((((((((((((.....))))......(((((.((((((.................))))))))))).........)))))))))...(..((((....))))..)))....... (-29.35 = -29.63 + 0.28)

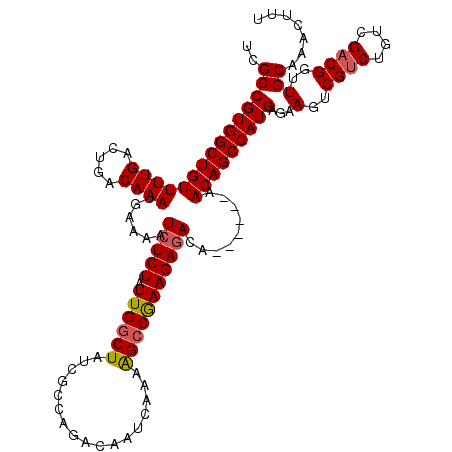
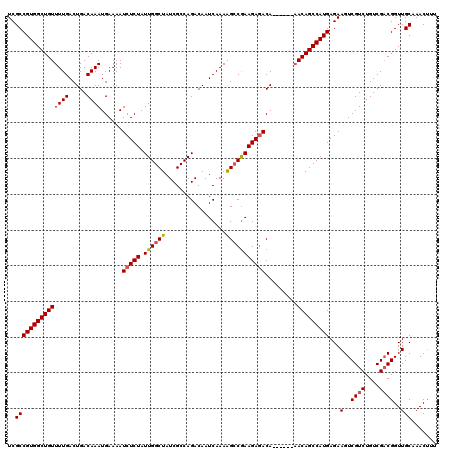
| Location | 21,051,683 – 21,051,797 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21051683 114 - 23771897 AAAGUUUGCAACCGUCGACAGACGACUUCUCAUGGCUGUU------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA ............((((((((((((.((......)).))))------)))).....((((((((((((.(((((....))))...(((...)))....).))))))))..))))..)))). ( -34.50) >DroSec_CAF1 106833 114 - 1 AAGGUUUGCAACCGGCGACAGACGACUUCUCAUGGCUGUG------UGUCUCUUCGUCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA ..(((.....)))(((((((((.((..((((.(((((((.------((((.....(((....))).....)))))))))))...))))...)).)))))).)))......((....)).. ( -33.60) >DroSim_CAF1 13046 114 - 1 AAAGUUUGCAACCGUCGACAGACGACUUCUCAUGGCUGUU------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA ............((((((((((((.((......)).))))------)))).....((((((((((((.(((((....))))...(((...)))....).))))))))..))))..)))). ( -34.50) >DroEre_CAF1 29232 120 - 1 AAAGUUUGCAACCGUCGACAGACGACUUCUCAUGGCUGUUUGUGUUUGUUUCUUUGGCCUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA .....((((...((((....))))........((((((((((((...(((((((((((..(((.........)))..))))).))))))...))).((.....)))))))))))..)))) ( -33.00) >DroYak_CAF1 307 114 - 1 AAAGUUUGCAACCGUCGACAGACGACUUCUCAUGGCUGUU------UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA ............((((((((((((.((......)).))))------)))).....((((((((((((.(((((....))))...(((...)))....).))))))))..))))..)))). ( -34.50) >consensus AAAGUUUGCAACCGUCGACAGACGACUUCUCAUGGCUGUU______UGUCUCUUCGGCUUUUGAUUGUCUGGCGAUAGCCAAUAGAGAUUUUCAUUUGUCAGUCAAAACAGCCACGGCGA .....((((...((((....))))........(((((((........(((.....))).((((((((.(((((....))))...(((...)))....).)))))))))))))))..)))) (-30.14 = -30.38 + 0.24)

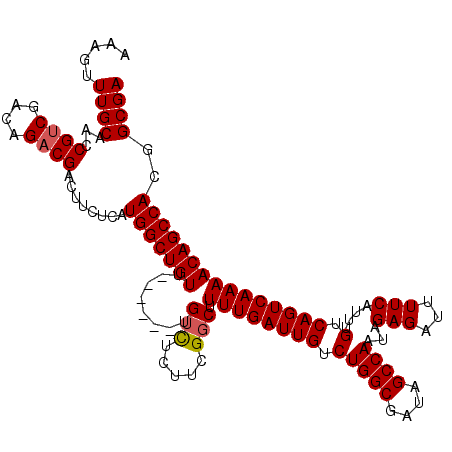
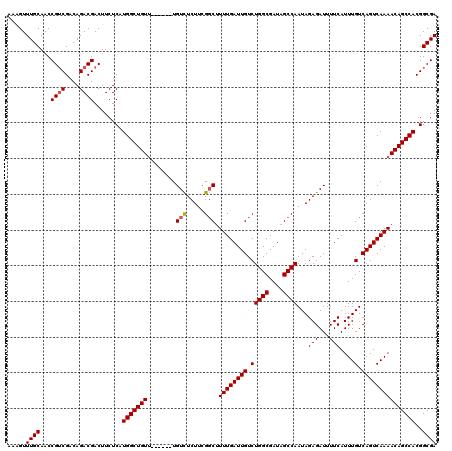
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:28 2006