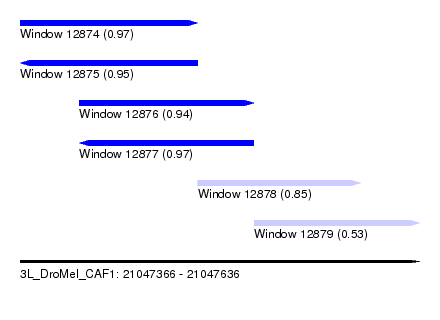
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,047,366 – 21,047,636 |
| Length | 270 |
| Max. P | 0.968846 |

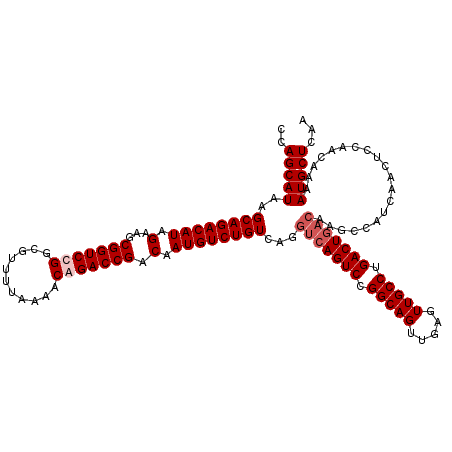
| Location | 21,047,366 – 21,047,486 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -40.30 |
| Energy contribution | -41.55 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047366 120 + 23771897 CCAGCAUAAGCAGACAUAGAAGCGGUCCGGCGUUUUAAAACAGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAA ..(((((..((((((((.(...(((((.(...........).))))).).))))))))...(((((((.(((((.....))))).)))))))....................)))))... ( -40.80) >DroSec_CAF1 102603 120 + 1 CCAGCAUAAGCAGACAUAGAAGCGGUCCGGCGUUUUAAAACGGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUACAACAAAAUGCUCAA ..(((((..((((((((.(...(((((((...........))))))).).))))))))...(((((((.(((((.....))))).)))))))....................)))))... ( -46.90) >DroSim_CAF1 8855 120 + 1 CCAGCAUAAGCAGACAUAGAAGCGGUCCGGCGUUUUAAAACGGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAA ..(((((..((((((((.(...(((((((...........))))))).).))))))))...(((((((.(((((.....))))).)))))))....................)))))... ( -46.90) >DroEre_CAF1 24650 115 + 1 CCAGCAUAAGCAGACAUAGAAACGGUCCGGCGUUUUAAAACAGACCGACAAUGUCUGUC-----AGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAA ..(((((....((((((.(...(((((.(...........).))))).).))))))(((-----((((.(((((.....))))).)))))))....................)))))... ( -35.70) >consensus CCAGCAUAAGCAGACAUAGAAGCGGUCCGGCGUUUUAAAACAGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAA ..(((((..((((((((.(...(((((((...........))))))).).))))))))...(((((((.(((((.....))))).)))))))....................)))))... (-40.30 = -41.55 + 1.25)

| Location | 21,047,366 – 21,047,486 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -38.72 |
| Energy contribution | -39.47 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047366 120 - 23771897 UUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCUGUUUUAAAACGCCGGACCGCUUCUAUGUCUGCUUAUGCUGG .((((((...(((..((((.....))).(((((((.((((.......)))).))))))))..)))(((((.(.(((((((...........))))))))....)))))))))))...... ( -42.10) >DroSec_CAF1 102603 120 - 1 UUGAGCAUUUUGUUGUAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCGUUUUAAAACGCCGGACCGCUUCUAUGUCUGCUUAUGCUGG ...(((((...((..(.....)..))..(((((((.((((.......)))).)))))))....(((((((.(.(((((((...........))))))))....)))))))...))))).. ( -42.80) >DroSim_CAF1 8855 120 - 1 UUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCGUUUUAAAACGCCGGACCGCUUCUAUGUCUGCUUAUGCUGG .((((((...(((..((((.....))).(((((((.((((.......)))).))))))))..)))(((((.(.(((((((...........))))))))....)))))))))))...... ( -44.30) >DroEre_CAF1 24650 115 - 1 UUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACU-----GACAGACAUUGUCGGUCUGUUUUAAAACGCCGGACCGUUUCUAUGUCUGCUUAUGCUGG ...(((((...((..((((.....))))(((((((.((((.......)))).))))-----)))((((((.(.(((((((...........)))))))...).))))))))..))))).. ( -40.80) >consensus UUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCGUUUUAAAACGCCGGACCGCUUCUAUGUCUGCUUAUGCUGG ...(((((.......((((.....))))(((((((.((((.......)))).)))))))....(((((((.(.(((((((...........)))))))...).)))))))...))))).. (-38.72 = -39.47 + 0.75)

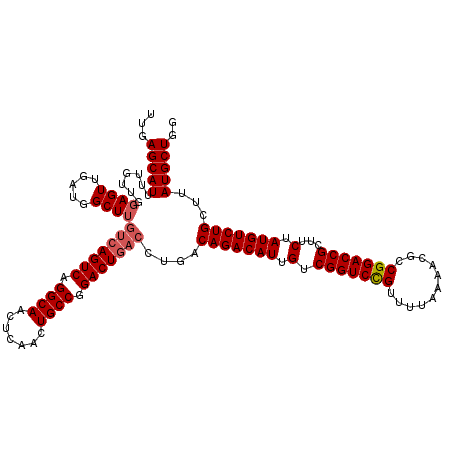
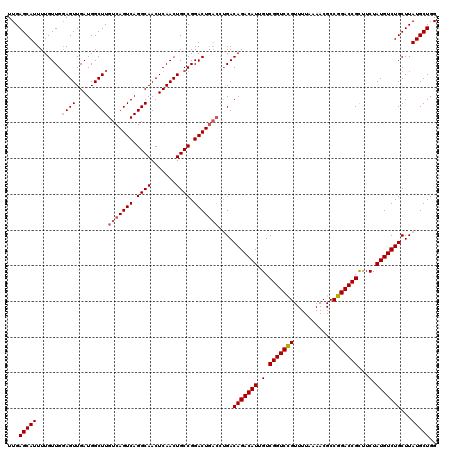
| Location | 21,047,406 – 21,047,524 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -27.30 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047406 118 + 23771897 CAGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAACACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAA-- ..((((((((.....)))).))))..((((.(.(((((((((..((.((....)).)).))))).)))).....((((..............)))).).))))...............-- ( -31.04) >DroSec_CAF1 102643 120 + 1 CGGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUACAACAAAAUGCUCAACACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAACA .((...((((.....))))..(((((((.(((((.....))))).)))))))...))...........................(((((........))))).................. ( -31.40) >DroSim_CAF1 8895 120 + 1 CGGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAACACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAACA .(((..((((.....))))..(((((((.(((((.....))))).)))))))...........)))..................(((((........))))).................. ( -32.60) >DroEre_CAF1 24690 111 + 1 CAGACCGACAAUGUCUGUC-----AGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAACACCUCGCCAAAGAGCCGGCGGAAAACGAAA--UAAAA-- .....((.....(..((((-----((((.(((((.....))))).))))))))..)............................(((((........)))))....))...--.....-- ( -29.00) >consensus CAGACCGACAAUGUCUGUCAGGUCAGUCCGGCAGUUGAGUUGCCUGACUGACAAGCCAUCAACUCCAACAAAAUGCUCAACACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAA__ ..(((.(((...))).)))..(((((((.(((((.....))))).)))))))................................(((((........))))).................. (-27.30 = -28.05 + 0.75)


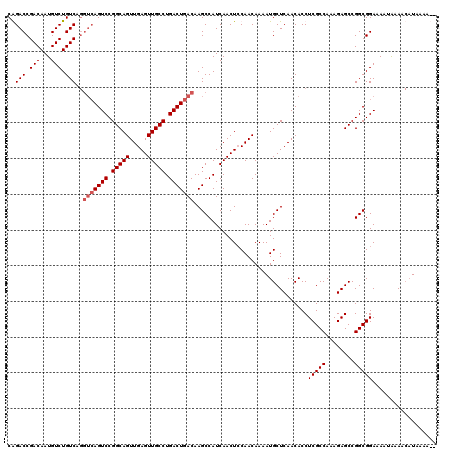
| Location | 21,047,406 – 21,047,524 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -43.25 |
| Consensus MFE | -37.10 |
| Energy contribution | -38.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047406 118 - 23771897 --UUUUAUGUUUUAUUUUCCGCCGGCUCUUUGGCGAGGUGUUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCUG --..................((((...(((..((((((((.....))))))))..)))....))))..(((((((.((((.......)))).)))))))(((((((...))))))).... ( -43.40) >DroSec_CAF1 102643 120 - 1 UGUUUUAUGUUUUAUUUUCCGCCGGCUCUUUGGCGAGGUGUUGAGCAUUUUGUUGUAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCG ......((((((.((.(..((((((....))))))..).)).))))))......((((((((..(((((.....)))))...))))))))(((((((((.((.....))..))))))))) ( -41.50) >DroSim_CAF1 8895 120 - 1 UGUUUUAUGUUUUAUUUUCCGCCGGCUCUUUGGCGAGGUGUUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCG ....................((((...(((..((((((((.....))))))))..)))....))))..(((((((.((((.......)))).)))))))(((((((...))))))).... ( -43.40) >DroEre_CAF1 24690 111 - 1 --UUUUA--UUUCGUUUUCCGCCGGCUCUUUGGCGAGGUGUUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACU-----GACAGACAUUGUCGGUCUG --.....--...........((((((.(((..((((((((.....))))))))..)))..((((..(((((((((.((((.......)))).))))-----))))).))))))))))... ( -44.70) >consensus __UUUUAUGUUUUAUUUUCCGCCGGCUCUUUGGCGAGGUGUUGAGCAUUUUGUUGGAGUUGAUGGCUUGUCAGUCAGGCAACUCAACUGCCGGACUGACCUGACAGACAUUGUCGGUCCG ....................((((...(((..((((((((.....))))))))..)))....))))..(((((((.((((.......)))).)))))))..(((.(((...))).))).. (-37.10 = -38.10 + 1.00)



| Location | 21,047,486 – 21,047,596 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.39 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047486 110 + 23771897 CACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAA-------CAUAAAAAGCUGGGG-AAAAACAAGAGCCACUGCAGAGACUUUCUUCGCAGACCAGCUUU--UUUCGAUGCAUCA ....(((((........)))))................-------(..((((((((((((-...........)).((((..((.....))..)))).))))))))--))..)........ ( -28.70) >DroSec_CAF1 102723 119 + 1 CACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAACAUGCAACAUAAAAAGCUGGGG-AAAAUCAAGAGCCACUGCAGAGACUUUCCUCGCAGACCAGCUUUUUUUUCGAUGCAUCA ....(((((........))))).................(((((.(..((((((((((((-...........)).((((.(((......))))))).))))))))))....).))))).. ( -33.30) >DroSim_CAF1 8975 118 + 1 CACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAACAUCAAACAUAAAAAGCUGGGG-AAAAACAAGAGCCACUGCAGAGACUUUCCUUGCAGACCAGCUUU-UUUUCGAUGCAUCA ....(((((........)))))................((((......((((((((((((-...........)).((((((.(.....).)))))).))))))))-))...))))..... ( -30.30) >DroEre_CAF1 24765 107 + 1 CACCUCGCCAAAGAGCCGGCGGAAAACGAAA--UAAAA-------CACA--AAACUGUGGAAAAAACAAGAGCCACUGCAGAGACUUUCCUCGCAGACUCGCUUU--UUUCGAUGCAUCA ....(((((........))))).....((..--.....-------((((--....))))(((((((...(((...((((.(((......))))))).)))..)))--))))......)). ( -25.70) >consensus CACCUCGCCAAAGAGCCGGCGGAAAAUAAAACAUAAAA_______CAUAAAAAGCUGGGG_AAAAACAAGAGCCACUGCAGAGACUUUCCUCGCAGACCAGCUUU__UUUCGAUGCAUCA ....(((((........)))))............................((((((((((............)).((((.(((......))))))).))))))))............... (-20.04 = -20.48 + 0.44)

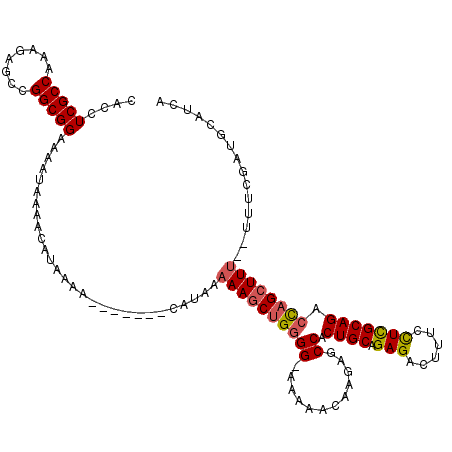

| Location | 21,047,524 – 21,047,636 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21047524 112 + 23771897 -----CAUAAAAAGCUGGGG-AAAAACAAGAGCCACUGCAGAGACUUUCUUCGCAGACCAGCUUU--UUUCGAUGCAUCAUCGCAUAAUCAAAGAGUCAUAAAAACCAAUAAUCAUUAUU -----...((((((((((((-...........)).((((..((.....))..)))).))))))))--))...((((......)))).................................. ( -24.80) >DroSec_CAF1 102763 119 + 1 UGCAACAUAAAAAGCUGGGG-AAAAUCAAGAGCCACUGCAGAGACUUUCCUCGCAGACCAGCUUUUUUUUCGAUGCAUCAUCGCAUAAUCAAAGAGUCAUAAAAACCAAUAAUCAUUAUU (((.....((((((((((((-...........)).((((.(((......))))))).))))))))))....((((...)))))))................................... ( -26.60) >DroSim_CAF1 9015 118 + 1 UCAAACAUAAAAAGCUGGGG-AAAAACAAGAGCCACUGCAGAGACUUUCCUUGCAGACCAGCUUU-UUUUCGAUGCAUCAUCGCAUAAUCAAAGAGUCAUAAAAACCAAUAAUCAUUAUU ........((((((((((((-...........)).((((((.(.....).)))))).))))))))-))....((((......)))).................................. ( -23.90) >DroEre_CAF1 24801 111 + 1 -----CACA--AAACUGUGGAAAAAACAAGAGCCACUGCAGAGACUUUCCUCGCAGACUCGCUUU--UUUCGAUGCAUCAUCGCAUAAUCAAAGAGUCAUAAAAACCAAUAAUCAUUAUU -----....--......(((.........(((...((((.(((......))))))).)))(((((--((...((((......))))....)))))))........)))............ ( -21.30) >consensus _____CAUAAAAAGCUGGGG_AAAAACAAGAGCCACUGCAGAGACUUUCCUCGCAGACCAGCUUU__UUUCGAUGCAUCAUCGCAUAAUCAAAGAGUCAUAAAAACCAAUAAUCAUUAUU ..........((((((((((............)).((((.(((......))))))).)))))))).......((((......)))).................................. (-16.44 = -16.88 + 0.44)

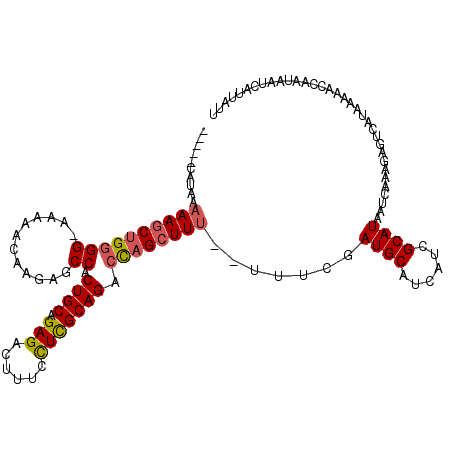

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:16 2006