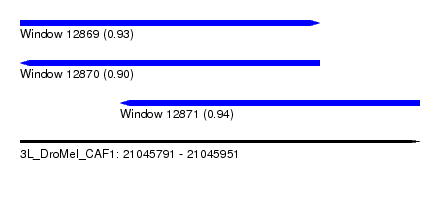
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,045,791 – 21,045,951 |
| Length | 160 |
| Max. P | 0.942980 |

| Location | 21,045,791 – 21,045,911 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.01 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21045791 120 + 23771897 ACUCAUUUCCCAGUCCACAGAAAUGCACGACCACUACUGAGGAGGAAAACUCCCACUUCGUUGAAAACUAUUUCCACCCGCUGAUGACACAUUACUGUCACUCAGCGUUAUGAGCUAAAU .(((((.............((((((..((((......((.((((.....))))))....)))).....))))))....(((((((((((......))))).))))))..)))))...... ( -27.80) >DroSec_CAF1 101072 120 + 1 ACUCAUUUCUCAGUCCACAGAAAUGCACGGCCUCUGCUGAGGAGGAAAACUCCCACUUCGUUAGAAACUAUUUCCACCCGCUGAUGACACAUUACUGUCACUCAGCGUUAUGAGCUAAAU .(((((((((((((((.((....))...)).....))))))))(((((........(((....)))....)))))...(((((((((((......))))).))))))..)))))...... ( -31.00) >DroSim_CAF1 7311 120 + 1 ACUCAUUUCUCAGUACACAGAAAUGCACGGCCACUGCUGAGGAGGAAAACUCCCACUUCGUUAGAAACUAUUUCCACCCGCUGAUGACACAUUACUGUCACUCAGCGUUAUGAGCUAAAU .((((((((((((((..((....))...(....))))))))))(((((........(((....)))....)))))...(((((((((((......))))).))))))..)))))...... ( -30.60) >DroEre_CAF1 23109 120 + 1 ACUCAUUUCCCACUCCGUGGAAAUGCACGAACACUGCUGAGGAGGAAAACUCCCACUUCGUUAGAAACUAUUUCCACCCGCUGAUGACACAUUACUGUCACUCAGCGUUAUGAGCUAAAU .(((((..........(((((((((....(((...(.((.((((.....)))))))...)))......))))))))).(((((((((((......))))).))))))..)))))...... ( -35.60) >consensus ACUCAUUUCCCAGUCCACAGAAAUGCACGACCACUGCUGAGGAGGAAAACUCCCACUUCGUUAGAAACUAUUUCCACCCGCUGAUGACACAUUACUGUCACUCAGCGUUAUGAGCUAAAU .(((((.............((((((..........((.(((..(((....)))..))).)).......))))))....(((((((((((......))))).))))))..)))))...... (-26.19 = -26.01 + -0.19)

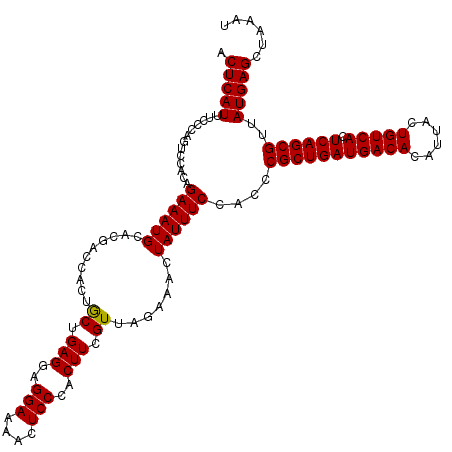
| Location | 21,045,791 – 21,045,911 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -34.67 |
| Energy contribution | -33.67 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21045791 120 - 23771897 AUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUUCAACGAAGUGGGAGUUUUCCUCCUCAGUAGUGGUCGUGCAUUUCUGUGGACUGGGAAAUGAGU ......(((((..((((((.(((((......))))))))))).((.((((....)))).))...))))).((((((((((.(((.((((......)))).))).)))..))))))).... ( -37.60) >DroSec_CAF1 101072 120 - 1 AUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCCUCAGCAGAGGCCGUGCAUUUCUGUGGACUGAGAAAUGAGU .....((((((..((((((.(((((......)))))))))))((.(((((...((((((......)))))).))))).))((.(((((.((...))...))))).)).......)))))) ( -36.80) >DroSim_CAF1 7311 120 - 1 AUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCCUCAGCAGUGGCCGUGCAUUUCUGUGUACUGAGAAAUGAGU .....((((((..((((((.(((((......)))))))))))(((.(.......(((....)))(((((((.....)))))..))..).)))((((((....))))))......)))))) ( -37.80) >DroEre_CAF1 23109 120 - 1 AUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCCUCAGCAGUGUUCGUGCAUUUCCACGGAGUGGGAAAUGAGU .....((((((..((((((.(((((......))))))))))).((((((((.((..(.((((..(((((((.....)))))..))..)))).).))))))))))..........)))))) ( -41.00) >consensus AUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCCUCAGCAGUGGCCGUGCAUUUCUGUGGACUGAGAAAUGAGU .....(((.....((((((.(((((......)))))))))))((.(((((...((((((......)))))).))))).))..)))..........(((((((........)))))))... (-34.67 = -33.67 + -1.00)


| Location | 21,045,831 – 21,045,951 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -39.47 |
| Energy contribution | -38.73 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21045831 120 - 23771897 AAUUGUGAGGGCGCGGUUUCUUGCUGGGCUGCAUCAAUAAAUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUUCAACGAAGUGGGAGUUUUCCUCC ..((((((((((...((((((.(((((((((.((......)).))))))....((((((.(((((......)))))))))))))))))))).)))))).))))....((((.....)))) ( -37.60) >DroSec_CAF1 101112 120 - 1 AAUUGUGAGGGCGCGGUUUCUUGCCGGGCUGCAUCAAUAAAUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCC ..((((((((((.((((.....)))).))).....((.....))..)))))))((((((.(((((......)))))))))))((.(((((...((((((......)))))).))))).)) ( -39.40) >DroSim_CAF1 7351 119 - 1 AAUUGUGAGGGCGCGGUUUC-UGCCGGGCUGCAUCAAUAAAUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCC ..((((..(((.((.(((((-..((((((((.((......)).))))))....((((((.(((((......)))))))))))))..))))).)).))).))))....((((.....)))) ( -41.30) >DroEre_CAF1 23149 120 - 1 AAUUGUGAGGGCGCGGCUUCUUGCCGUGCUGCAUCAAUAAAUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCC ..(((((((((((((((.....)))))))).....((.....))..)))))))((((((.(((((......)))))))))))((.(((((...((((((......)))))).))))).)) ( -43.70) >consensus AAUUGUGAGGGCGCGGUUUCUUGCCGGGCUGCAUCAAUAAAUUUAGCUCAUAACGCUGAGUGACAGUAAUGUGUCAUCAGCGGGUGGAAAUAGUUUCUAACGAAGUGGGAGUUUUCCUCC ..((((((((((.((((.....)))).))).....((.....))..)))))))((((((.(((((......)))))))))))((.(((((...((((((......)))))).))))).)) (-39.47 = -38.73 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:08 2006