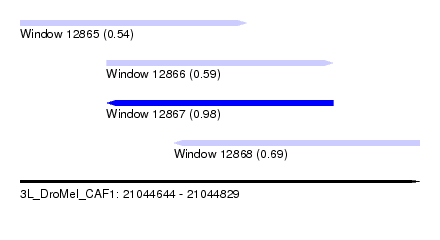
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,044,644 – 21,044,829 |
| Length | 185 |
| Max. P | 0.979082 |

| Location | 21,044,644 – 21,044,749 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -24.01 |
| Energy contribution | -25.57 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044644 105 + 23771897 UUUGCAUUGCAAAAGGAAAGUGACAAAAAGGCGAGAGGAAAAU--AUAAGGAGACAGUGGCACUGGCAGUGGC-------------GAUCUGCGAUCACUGAUCACAAGUCGGCGAUACG .(((((((((...((....((.((.......(....)......--....(....).)).)).)).)))))..(-------------((..((.((((...)))).))..))))))).... ( -23.20) >DroSec_CAF1 99880 120 + 1 UUUGCAUUGCAAAAGGAAAGUGACAAAAAGGCGAGAGGAAAAUGUAUAAGGAGACAGUGGCAGUGGCAGUGGCAGUGCCACAUGGGGAACUGCGAUCACUGAUCACAAGUCGCCGAUACG ((((((((..........)))).))))..(((((...............(....).(((((((((...((.(((((.((......)).))))).))))))).))))...)))))...... ( -35.40) >DroSim_CAF1 6128 120 + 1 UUUGCAUUGCAAAAGGAAAGUGACAAAAAGGCGAGAGGAAAAUAUACAAGGAGACAGUGGCAGUGGCAGUGGCAGUGCCACAUGGCGAUCUGCGAUCACUGAUCACAAGUCGCCGAUACG ((((((((..........)))).))))..(((((...............(....).((((((((((..(..(...((((....))))..)..)..)))))).))))...)))))...... ( -36.20) >DroEre_CAF1 22076 111 + 1 UUUGCAUUGCAAAAGGAAAGUGACAAAAAGGCGACAGAAAAAUG--UAAGGAGACAGUGGCAGUGGCAGUGGCAGUGU-------UGAUCAGCGAUCACUGAUCACAACUCGUCGAUACG .(((((((((.......................(((......))--)..(....)....))))).))))(((((((..-------(((((((......)))))))..))).))))..... ( -25.60) >consensus UUUGCAUUGCAAAAGGAAAGUGACAAAAAGGCGAGAGGAAAAUG_AUAAGGAGACAGUGGCAGUGGCAGUGGCAGUGC________GAUCUGCGAUCACUGAUCACAAGUCGCCGAUACG ((((((((..........)))).))))..(((((...............(....).((((((((((..((((...((((....))))..))))..)))))).))))...)))))...... (-24.01 = -25.57 + 1.56)


| Location | 21,044,684 – 21,044,789 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -29.13 |
| Energy contribution | -29.69 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044684 105 + 23771897 AAU--AUAAGGAGACAGUGGCACUGGCAGUGGC-------------GAUCUGCGAUCACUGAUCACAAGUCGGCGAUACGGCAAGGAUGCCAUAAUUGCGUGUGAGUUUGGCCACUCGUU ...--....(....)(((((((((.(((....(-------------((..((.((((...)))).))..)))(((((..((((....))))...))))).))).)))...)))))).... ( -34.40) >DroSec_CAF1 99920 120 + 1 AAUGUAUAAGGAGACAGUGGCAGUGGCAGUGGCAGUGCCACAUGGGGAACUGCGAUCACUGAUCACAAGUCGCCGAUACGGCAAGGAUGCCAUAAUUGCGUGUGAGUUUGGCCACUCGUU .........(....)((((((.((((((((((((((.((......)).)))))...))))).))))((.((((((.((.((((....)))).))....)).)))).))..)))))).... ( -41.70) >DroSim_CAF1 6168 120 + 1 AAUAUACAAGGAGACAGUGGCAGUGGCAGUGGCAGUGCCACAUGGCGAUCUGCGAUCACUGAUCACAAGUCGCCGAUACGGCAAGGAUGCCAUAAUUGCGUGUGAGUUUGGCCACUCGUU ...((((..(....)....(((((....((((((.((((...((((((..((.((((...)))).))..))))))....))))....)))))).)))))))))((((......))))... ( -43.40) >DroEre_CAF1 22116 111 + 1 AAUG--UAAGGAGACAGUGGCAGUGGCAGUGGCAGUGU-------UGAUCAGCGAUCACUGAUCACAACUCGUCGAUACGGCAAGGAUGCCAUAAUUGCGUGUGAGUUUGGCCAUUCGCU .(((--((((....).((((((((((((((((...(((-------(....)))).)))))).))))..((.((((...)))).))..))))))..))))))((((((......)))))). ( -35.40) >consensus AAUG_AUAAGGAGACAGUGGCAGUGGCAGUGGCAGUGC________GAUCUGCGAUCACUGAUCACAAGUCGCCGAUACGGCAAGGAUGCCAUAAUUGCGUGUGAGUUUGGCCACUCGUU .........(....)((((((.....(((((((((..............))))...))))).(((((...((.((((..((((....))))...))))))))))).....)))))).... (-29.13 = -29.69 + 0.56)



| Location | 21,044,684 – 21,044,789 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -28.75 |
| Energy contribution | -30.07 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044684 105 - 23771897 AACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGCCGACUUGUGAUCAGUGAUCGCAGAUC-------------GCCACUGCCAGUGCCACUGUCUCCUUAU--AUU ....(((((((..........((...(((((((....)))))))..))(((.((((((((...)))))))).))-------------).........).))))))..........--... ( -31.70) >DroSec_CAF1 99920 120 - 1 AACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGUUCCCCAUGUGGCACUGCCACUGCCACUGCCACUGUCUCCUUAUACAUU ....((((((.....((((((((...(((((((....)))))))...)))...))))).(((((...(((((.((......)).)))))))))).....))))))............... ( -41.70) >DroSim_CAF1 6168 120 - 1 AACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGAUCGCCAUGUGGCACUGCCACUGCCACUGCCACUGUCUCCUUGUAUAUU ....((((((................(((((((....)))))))..(((((.((((((((...)))))))).)))))..((((((.......)))))).))))))............... ( -45.40) >DroEre_CAF1 22116 111 - 1 AGCGAAUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGACGAGUUGUGAUCAGUGAUCGCUGAUCA-------ACACUGCCACUGCCACUGCCACUGUCUCCUUA--CAUU .(((..((((..(((((...((....(((((((....))))))).))..))))).((((((((....))))))))-------.....)))).)))...................--.... ( -31.10) >consensus AACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGAUC________GCACUGCCACUGCCACUGCCACUGUCUCCUUAU_CAUU .((.((((((.....((((((((...(((((((....)))))))...)))...))))).(((((...((((..............))))))))).....))))))))............. (-28.75 = -30.07 + 1.31)



| Location | 21,044,715 – 21,044,829 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044715 114 - 23771897 UCGUUGGUGCUAGCCUACAUACAUAACAAUUAGGCAACAAAACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGCCGACUUGUGAUCAGUGAUCGCAGAUC------ ..((((((((..(((((.............)))))........((((......))))....))...(((((((....)))))))..))))))((((((((...))))))))...------ ( -36.12) >DroSec_CAF1 99960 120 - 1 UCGCUGGUGCUAGCCUACAUACAUAACAAUUAGCCAACAAAACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGUUCCCCAUG ((((((((((.................................((((......))))............((((....)))))))))))))).((((((((...))))))))......... ( -32.70) >DroSim_CAF1 6208 120 - 1 UCGGCGGUGCUAGCCUACAUACAUAACAAUUAGCCAACAAAACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGAUCGCCAUG ...((((((((((................))))).........((((......)))))).)))...(((((((....)))))))..(((((.((((((((...)))))))).)))))... ( -38.39) >DroEre_CAF1 22152 115 - 1 UCGGUGGUGCUAGCCUACAUACAUAACAAUUAGCCAACGGAGCGAAUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGACGAGUUGUGAUCAGUGAUCGCUGAUCA----- ...((((.(....))))).......((((((.((((..........))))..........((....(((((((....)))))))....))))))))(((((((....))))))).----- ( -30.60) >consensus UCGGUGGUGCUAGCCUACAUACAUAACAAUUAGCCAACAAAACGAGUGGCCAAACUCACACGCAAUUAUGGCAUCCUUGCCGUAUCGGCGACUUGUGAUCAGUGAUCGCAGAUC______ .....(((....))).................(((........((((......)))).........(((((((....)))))))..)))((.((((((((...)))))))).))...... (-26.60 = -27.10 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:02 2006