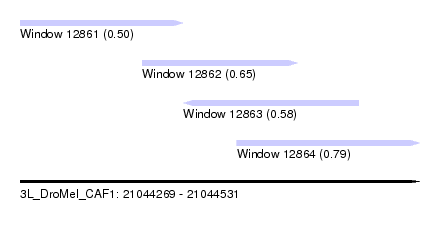
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,044,269 – 21,044,531 |
| Length | 262 |
| Max. P | 0.791290 |

| Location | 21,044,269 – 21,044,376 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044269 107 + 23771897 CGCGUGAUUUGUGAUGAGUGCAGCUAUGUUUUAGGUCUCUAAAUAUUCAUAAAUUCAGGCGCUCAUAAAGCUUUGGUUAAUAAAAAUCCUAAAACAGGAUUCGCAAA------------- .((((((((.((.((((((((...((((.(((((....)))))....)))).......))))))))...))...))))).....((((((.....)))))))))...------------- ( -25.20) >DroSec_CAF1 99504 107 + 1 CGCGUGAUUAGUGAUGAGUGCAGCUAAGUUGUAGGUCUGUAAAUAUUAAUAAAUUCGUGCGCUCAUAAAGCUUUGGUUAAUAAAAAUCCUAAAACAGGAUUCGCGAG------------- ((((((((((((.(((((((((......((((..((........))..)))).....)))))))))...)))..))))).....((((((.....))))))))))..------------- ( -25.20) >DroSim_CAF1 5752 107 + 1 CGCGUGAUUAGUGAUGAGUGCAGCUAAGUUGUAGGUCUGUAAAUAUUCAUAAAUUCAGGCGCUCAUCAAGCUUUGGCUAAUAAAAAUCCUAAAACAGGAUUCGCGAG------------- ((((.......(((((((((((((...)))))..(((((................)))))))))))))(((....)))......((((((.....))))))))))..------------- ( -29.49) >DroEre_CAF1 21678 120 + 1 UGCGUGAUUUGUGAUCAGUGCAUCCAUGUUUAAGGUCUGUAAAUAUUCAUAAAUUCAGUCGCUCAUAAAGCUUUGGCUCAUAAAAAUCCUAAAACAGGAUUUGCGAACCCUGGUGCUGUU .(((.(((((((((....((((.((........))..)))).....)))))))))(((((((......(((....))).....(((((((.....)))))))))))...))).))).... ( -26.60) >consensus CGCGUGAUUAGUGAUGAGUGCAGCUAAGUUGUAGGUCUGUAAAUAUUCAUAAAUUCAGGCGCUCAUAAAGCUUUGGCUAAUAAAAAUCCUAAAACAGGAUUCGCGAA_____________ .((.((((((((((....(((((((........)).))))).....))))))).))).))((......(((....)))......((((((.....)))))).))................ (-18.95 = -19.83 + 0.87)

| Location | 21,044,349 – 21,044,451 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -23.16 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044349 102 + 23771897 UAAAAAUCCUAAAACAGGAUUCGCAAA------------------GCAGAGAAACUUUCCUCGAAAUCUGCAUAUAAGCCGCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUC .....(((((.....)))))((((...------------------(((((................)))))........(((((((((........).))))))))........)))).. ( -25.69) >DroSec_CAF1 99584 102 + 1 UAAAAAUCCUAAAACAGGAUUCGCGAG------------------CCAGAGAAACUUUCCUCGAAAUCUGCAUAUAAGCCGCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUC .....(((((.....)))))((((..(------------------((((((........))).................(((((((((........).))))))))..)).)).)))).. ( -24.70) >DroSim_CAF1 5832 102 + 1 UAAAAAUCCUAAAACAGGAUUCGCGAG------------------CCAGAGAAACUUUCCUCGAAAUCUGCAUAUAAGCCGCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUC .....(((((.....)))))((((..(------------------((((((........))).................(((((((((........).))))))))..)).)).)))).. ( -24.70) >DroEre_CAF1 21758 120 + 1 UAAAAAUCCUAAAACAGGAUUUGCGAACCCUGGUGCUGUUUUCCACCAGAGAAACUUUCCUCGAAAUCUGCAUAUAAGCCGCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUC ...(((((((.....)))))))((.....((((((........))))))..........((((......((......))(((((((((........).))))))))..))))..)).... ( -29.90) >consensus UAAAAAUCCUAAAACAGGAUUCGCGAA__________________CCAGAGAAACUUUCCUCGAAAUCUGCAUAUAAGCCGCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUC .....(((((.....)))))((((........................(((........)))......(((.(((....(((((((((........).)))))))).))).))))))).. (-23.16 = -22.98 + -0.19)

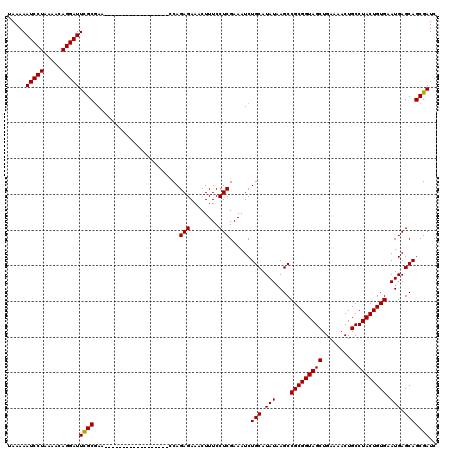
| Location | 21,044,376 – 21,044,491 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.45 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -29.12 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044376 115 - 23771897 CUCCUCCAGUCACCUGAAUUUUCCUUCUCCUGCCCACGAAGAUCGCUGCUCAUUCACAGUAGGCAGUUUUCAGCUACCGCGGCUUAUAUGCAGAUUUCGAGGAAAGUUUCUCUGC----- ......(((......(((((((((((...((((.......((..(((((..((.....))..)))))..))((((.....)))).....)))).....)))))))))))..))).----- ( -27.50) >DroSec_CAF1 99611 115 - 1 CUCCUCCAGUCACCUGAAUUUUCCUUCUCCUGCCCACGAAGAUCGCUGCUCAUUCACAGUAGGCAGUUUUCAGCUACCGCGGCUUAUAUGCAGAUUUCGAGGAAAGUUUCUCUGG----- .....((((......(((((((((((...((((.......((..(((((..((.....))..)))))..))((((.....)))).....)))).....)))))))))))..))))----- ( -30.20) >DroSim_CAF1 5859 115 - 1 CUCCUCCAGUCACCUGAAUUUUCCUUCUCCUGCCCACGAAGAUCGCUGCUCAUUCACAGUAGGCAGUUUUCAGCUACCGCGGCUUAUAUGCAGAUUUCGAGGAAAGUUUCUCUGG----- .....((((......(((((((((((...((((.......((..(((((..((.....))..)))))..))((((.....)))).....)))).....)))))))))))..))))----- ( -30.20) >DroEre_CAF1 21798 120 - 1 CUCCUCCAGUCACCUGAAUUUUCCUUCUCCUGCCCACGAAGAUCGCUGCUCAUUCACAGUAGGCAGUUUUCAGCUACCGCGGCUUAUAUGCAGAUUUCGAGGAAAGUUUCUCUGGUGGAA .(((.((((......(((((((((((...((((.......((..(((((..((.....))..)))))..))((((.....)))).....)))).....)))))))))))..)))).))). ( -36.50) >consensus CUCCUCCAGUCACCUGAAUUUUCCUUCUCCUGCCCACGAAGAUCGCUGCUCAUUCACAGUAGGCAGUUUUCAGCUACCGCGGCUUAUAUGCAGAUUUCGAGGAAAGUUUCUCUGG_____ .....((((......(((((((((((...((((.......((..(((((..((.....))..)))))..))((((.....)))).....)))).....)))))))))))..))))..... (-29.12 = -29.38 + 0.25)

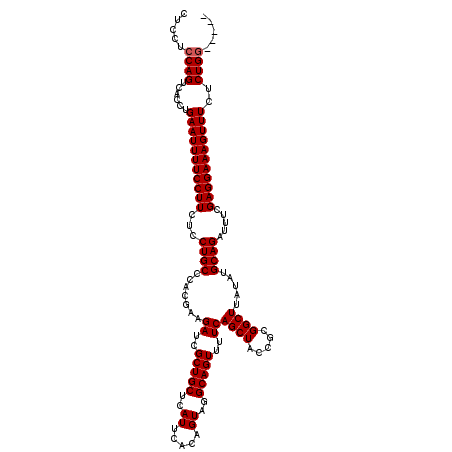
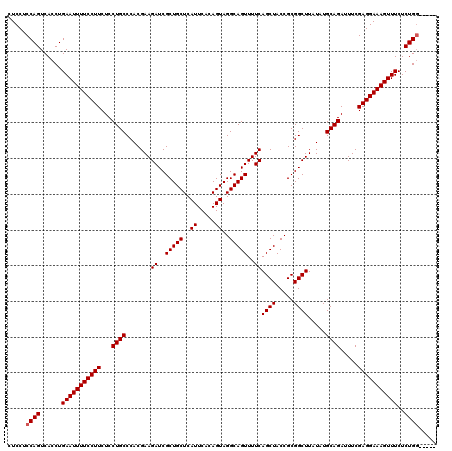
| Location | 21,044,411 – 21,044,531 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -35.65 |
| Energy contribution | -38.90 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21044411 120 + 23771897 GCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUCUUCGUGGGCAGGAGAAGGAAAAUUCAGGUGACUGGAGGAGAUGGGAUGGAGUGGGGGAUAUUAGAGCCAGGUUGCCGCAU (((((((((.....((((((((...((.((.....)).))...))))))))...........(((((....))))).......((....((((.....))))....)).))))))))).. ( -39.40) >DroSec_CAF1 99646 120 + 1 GCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUCUUCGUGGGCAGGAGAAGGAAAAUUCAGGUGACUGGAGGAGAUGGGAUGGAGUGGGGGAUAUUAGAGCCAGGUUGCCGCAU (((((((((.....((((((((...((.((.....)).))...))))))))...........(((((....))))).......((....((((.....))))....)).))))))))).. ( -39.40) >DroSim_CAF1 5894 120 + 1 GCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUCUUCGUGGGCAGGAGAAGGAAAAUUCAGGUGACUGGAGGAGAUGGGAUGGAGUGGGGGAUAUUAGAGCCAGGUUGCCGCAU (((((((((.....((((((((...((.((.....)).))...))))))))...........(((((....))))).......((....((((.....))))....)).))))))))).. ( -39.40) >DroEre_CAF1 21838 120 + 1 GCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUCUUCGUGGGCAGGAGAAGGAAAAUUCAGGUGACUGGAGGAGAUGGGAUGGACUGGGGGAUAUUAAAGCCAGGUUGCCGCAU (((((((((.....((((((((...((.((.....)).))...))))))))...........(((((....)))))((...(..((((..(....)..))))..).)).))))))))).. ( -40.90) >consensus GCGGUAGCUGAAAACUGCCUACUGUGAAUGAGCAGCGAUCUUCGUGGGCAGGAGAAGGAAAAUUCAGGUGACUGGAGGAGAUGGGAUGGAGUGGGGGAUAUUAGAGCCAGGUUGCCGCAU (((((((((.....((((((((...((.((.....)).))...))))))))...........(((((....))))).......((....((((.....))))....)).))))))))).. (-35.65 = -38.90 + 3.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:58 2006