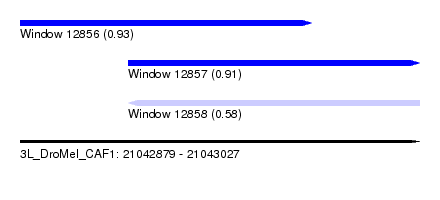
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,042,879 – 21,043,027 |
| Length | 148 |
| Max. P | 0.933427 |

| Location | 21,042,879 – 21,042,987 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.67 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21042879 108 + 23771897 UCAAAGCGAAGCGCCGUUCCAGUCUGGGAAUGGGAACCCAUG------------UACAUACAUAUCUACUUGUGUAUGUAUGGAGCUGCCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAG .....((((.((.(((((((......)))))))((.(((..(------------((((((((((......)))))))))))((((((......))))))...))).))))...)).)).. ( -40.80) >DroSec_CAF1 98138 117 + 1 UCAAAGCGAAGCGCCGUUCCAGUCUGGGAAUGGGAACCCAUGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG---CCUGGAGCUCUUCAGGGAUUGUGAGUCAGCAG .....((((..(((.(((((.(((..(..((((....))))..)..)))....((((((((((((....))))))))))))((((---(.....)))))...))))).)))..)).)).. ( -40.50) >DroSim_CAF1 4367 117 + 1 UCAAAGCGAAGCGCCGUUCCAGUCUGGGAAUGGGAACCCAUGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG---CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAG .....((((..(((.(((((((((((((........)))).))))))).)).........(((((((((....)))))))))((.---((((((....))))))..)))))..)).)).. ( -40.90) >DroEre_CAF1 20277 113 + 1 CCAAAGCGAAGCGCCGUUCCAGUCUGGGAAUGGGAUCCCAUGGUUGGGUACAUACUUGUACAUGUGUA----UGUAUGUACGGAG---CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAC .....((((.((.(((((((......)))))))((((((..((((..((((((((..((((....)))----)))))))))..))---))((((....))))))))))))...)).)).. ( -43.60) >consensus UCAAAGCGAAGCGCCGUUCCAGUCUGGGAAUGGGAACCCAUGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG___CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAG .....((((..(((.(((((((((((((........)))).)))))).........(((((((((........)))))))))......((((((....))))))))).)))..)).)).. (-32.30 = -32.67 + 0.38)

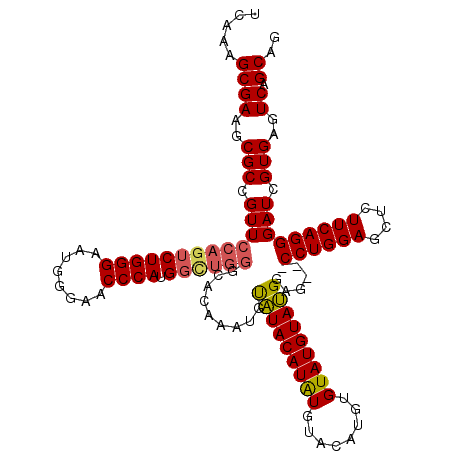
| Location | 21,042,919 – 21,043,027 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -25.27 |
| Energy contribution | -24.78 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21042919 108 + 23771897 UG------------UACAUACAUAUCUACUUGUGUAUGUAUGGAGCUGCCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAGAGUUUUGGCUAAGAAAUCUGCUAAAAAUAUAAGCCUCGAU .(------------((((((((((......)))))))))))(..(((.((((((....))))))((.(....)))((((((.((((.....)))).)))))).........)))..)... ( -35.40) >DroSec_CAF1 98178 117 + 1 UGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG---CCUGGAGCUCUUCAGGGAUUGUGAGUCAGCAGAGUUUUGGCUAAGAAAUCUGCUAAAAAUAUAAGCCUCGAU .((((((.(((((.......(((((((((....)))))))))...---((((((....))))))..)))))..))((((((.((((.....)))).)))))).........))))..... ( -36.20) >DroSim_CAF1 4407 117 + 1 UGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG---CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAGAGUUUUGGCUAAGAAAUCUGCUAAAAAUAUAAGCCUCGAU .((((.(((.((........(((((((((....)))))))))((.---((((((....))))))..)).)).)))((((((.((((.....)))).)))))).........))))..... ( -35.60) >DroEre_CAF1 20317 113 + 1 UGGUUGGGUACAUACUUGUACAUGUGUA----UGUAUGUACGGAG---CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCACAGUUUUGGCUAAGAAAUCUGCUAAAAAUAUAAGCCUCGAU .((((..(((((....))))).(((((.----((....(((((..---((((((....))))))..)))))...))))))).(((((((..(....)..))))))).....))))..... ( -32.60) >consensus UGGCUGGGCACAAAUGUAUACAUAUGUACAUGUGUAUGUAUGGAG___CCUGGAGCUCUUCAGGGAUCGUGAGUCAGCAGAGUUUUGGCUAAGAAAUCUGCUAAAAAUAUAAGCCUCGAU ................(((((((((........)))))))))(((...((((((....))))))...........((((((.((((.....)))).))))))............)))... (-25.27 = -24.78 + -0.50)

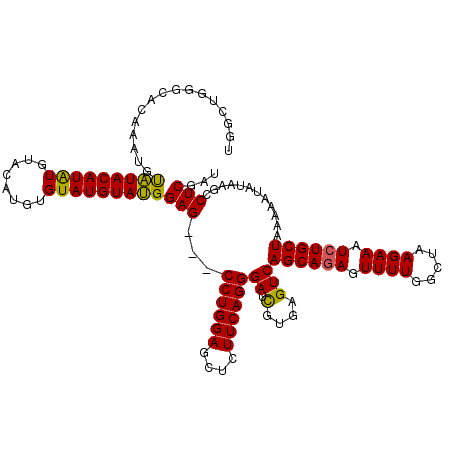
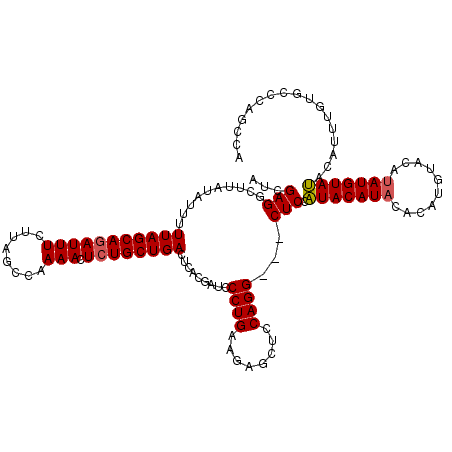
| Location | 21,042,919 – 21,043,027 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21042919 108 - 23771897 AUCGAGGCUUAUAUUUUUAGCAGAUUUCUUAGCCAAAACUCUGCUGACUCACGAUCCCUGAAGAGCUCCAGGCAGCUCCAUACAUACACAAGUAGAUAUGUAUGUA------------CA ...(..(((.......(((((((((((........))).)))))))).........((((........)))).)))..).((((((((..........))))))))------------.. ( -23.00) >DroSec_CAF1 98178 117 - 1 AUCGAGGCUUAUAUUUUUAGCAGAUUUCUUAGCCAAAACUCUGCUGACUCACAAUCCCUGAAGAGCUCCAGG---CUCCAUACAUACACAUGUACAUAUGUAUACAUUUGUGCCCAGCCA .....((((.......(((((((((((........))).))))))))..(((((........((((.....)---))).(((((((..........)))))))....)))))...)))). ( -26.10) >DroSim_CAF1 4407 117 - 1 AUCGAGGCUUAUAUUUUUAGCAGAUUUCUUAGCCAAAACUCUGCUGACUCACGAUCCCUGAAGAGCUCCAGG---CUCCAUACAUACACAUGUACAUAUGUAUACAUUUGUGCCCAGCCA .....((((.......(((((((((((........))).))))))))..(((((........((((.....)---))).(((((((..........)))))))....)))))...)))). ( -25.80) >DroEre_CAF1 20317 113 - 1 AUCGAGGCUUAUAUUUUUAGCAGAUUUCUUAGCCAAAACUGUGCUGACUCACGAUCCCUGAAGAGCUCCAGG---CUCCGUACAUACA----UACACAUGUACAAGUAUGUACCCAACCA ...(((.((....(((((((..((((..(((((((....)).))))).....)))).))))))).....)).---))).((((((((.----(((....)))...))))))))....... ( -25.40) >consensus AUCGAGGCUUAUAUUUUUAGCAGAUUUCUUAGCCAAAACUCUGCUGACUCACGAUCCCUGAAGAGCUCCAGG___CUCCAUACAUACACAUGUACAUAUGUAUACAUUUGUGCCCAGCCA ...(((..........(((((((((((........))).)))))))).........((((........))))...))).(((((((..........)))))))................. (-16.70 = -16.82 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:52 2006