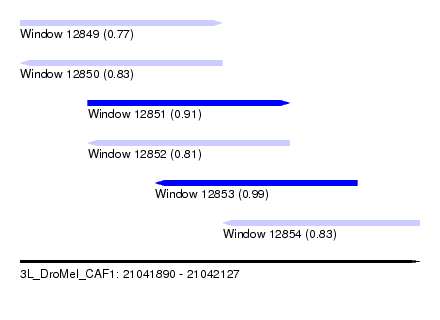
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,041,890 – 21,042,127 |
| Length | 237 |
| Max. P | 0.994242 |

| Location | 21,041,890 – 21,042,010 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -17.59 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21041890 120 + 23771897 AACUAUACUCAAUUACUCAACAAACAUAGUAACCUAGUUCAUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAGGCAACAAAUUCAUUUUCGCCUUUGUUGUU ......(((...(((((..........)))))...))).............((((......(((((....)))))))))..(((((.((((((...............)))))).))))) ( -21.06) >DroSec_CAF1 97150 120 + 1 AACUAUACUCAAUUACUCAACAAACAUAGUAACCUAGUUCAUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUU ......(((...(((((..........)))))...)))...............((((((((.(((((.....)))))(((......(((((............)))))))))))))))). ( -17.80) >DroSim_CAF1 3388 120 + 1 AACUAUACUCAAUUACUCAACAAACAUAGUAACCUAGUUCAUAAAAAAUAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUU ......(((...(((((..........)))))...)))...............((((((((.(((((.....)))))(((......(((((............)))))))))))))))). ( -17.80) >DroEre_CAF1 19207 120 + 1 AACCAUACUCAAUUACUCAACAAACAUAGUAACCUAGUUCAUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGUGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUU ..(((((((...(((((..........)))))...))).......................(((((....)))))))))...........(((((((((..(......)..))))))))) ( -17.40) >consensus AACUAUACUCAAUUACUCAACAAACAUAGUAACCUAGUUCAUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUU ......(((...(((((..........)))))...)))...............(((((((((((((....)))))..(((......(((((............)))))))))))))))). (-17.59 = -17.40 + -0.19)

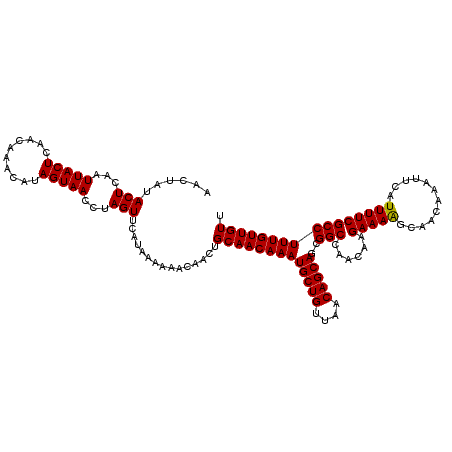

| Location | 21,041,890 – 21,042,010 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21041890 120 - 23771897 AACAACAAAGGCGAAAAUGAAUUUGUUGCCUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAUGAACUAGGUUACUAUGUUUGUUGAGUAAUUGAGUAUAGUU ..((((((((((.((...(((..........)))...)).)))..(((((....))))))))))))(((((((((...(..((((((....))).)))..).)))))))))......... ( -29.00) >DroSec_CAF1 97150 120 - 1 AACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAUGAACUAGGUUACUAUGUUUGUUGAGUAAUUGAGUAUAGUU ..((((((((((.((...(((..........)))...)).)))..(((((....))))))))))))(((((((((...(..((((((....))).)))..).)))))))))......... ( -29.00) >DroSim_CAF1 3388 120 - 1 AACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUAUUUUUUAUGAACUAGGUUACUAUGUUUGUUGAGUAAUUGAGUAUAGUU ..((((((((((.((...(((..........)))...)).)))..(((((....))))))))))))(((((((((...(..((((((....))).)))..).)))))))))......... ( -32.00) >DroEre_CAF1 19207 120 - 1 AACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCACUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAUGAACUAGGUUACUAUGUUUGUUGAGUAAUUGAGUAUGGUU (((((.((((((((.((.....)).)))))))).)))))(((((.(((((....))))).......(((((((((...(..((((((....))).)))..).)))))))))....))))) ( -29.20) >consensus AACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAUGAACUAGGUUACUAUGUUUGUUGAGUAAUUGAGUAUAGUU ..((((((((((.((...(((..........)))...)).)))..(((((....))))))))))))(((((((((...(..((((((....))).)))..).)))))))))......... (-29.94 = -29.75 + -0.19)

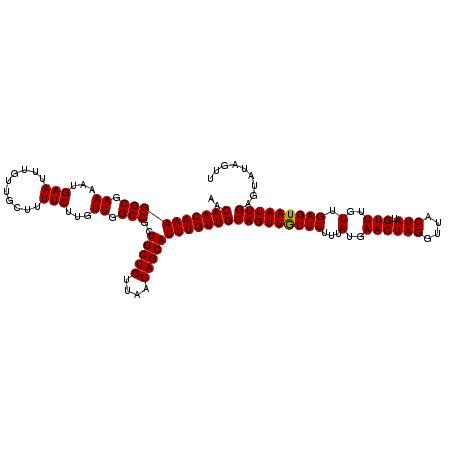

| Location | 21,041,930 – 21,042,050 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21041930 120 + 23771897 AUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAGGCAACAAAUUCAUUUUCGCCUUUGUUGUUUUGAUUUUUGCUUGGGUUUUUGUUGCUGGCGCUUUCCACG .............((((((((((((((.....)))))).((((((((((((((((((((..(......)..))))))))))...)))))).))))...))))))))(((......))).. ( -32.10) >DroSec_CAF1 97190 120 + 1 AUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUUUUGAUUUCUGCUUGGGUUUUUAUUGUUGGCGCUUUCCACG .............((......((((((.....))))))((((((((..(((((((((((..(......)..)))))))))))((..(((....)))..))..))))))))))........ ( -29.90) >DroSim_CAF1 3428 120 + 1 AUAAAAAAUAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUUUUGAUUUCUGCUUGGGUUUUUAUUGUUGGCGCUUUCCACG .............((......((((((.....))))))((((((((..(((((((((((..(......)..)))))))))))((..(((....)))..))..))))))))))........ ( -29.90) >DroEre_CAF1 19247 117 + 1 AUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGUGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUUUUGAUUUUUGCUUAAGUUUUU---GUUGGCGCUUUCCACG .............((..((..(((((....))))).))(((((((((((((((((((((..(......)..))))))))))).......((....))))))---))))))))........ ( -30.50) >consensus AUAAAAAACAACUGCAACAAAUGCUGUUAACAGCAGCGGCCAACAAGAAAAGCAACAAAUUCAUUUUCGCCUUUGUUGUUUUGAUUUCUGCUUGGGUUUUUAUUGUUGGCGCUUUCCACG .............((......((((((.....))))))((((((((..(((((((((((..(......)..)))))))))))(((((......)))))....))))))))))........ (-26.75 = -26.75 + 0.00)

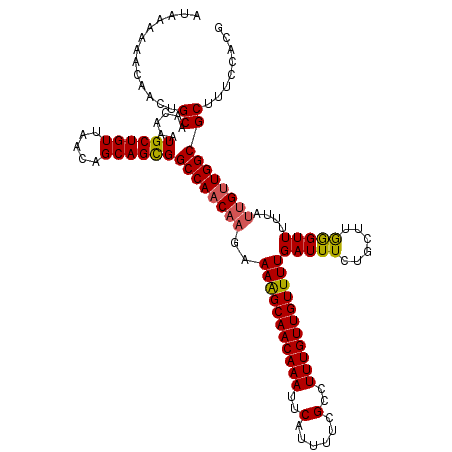
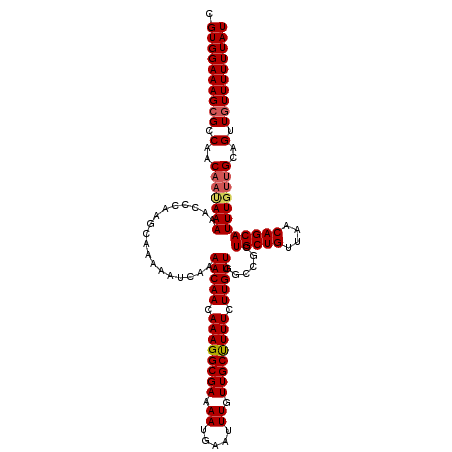
| Location | 21,041,930 – 21,042,050 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21041930 120 - 23771897 CGUGGAAAGCGCCAGCAACAAAAACCCAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCCUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAU .((((((((((.(.((((((((......(((.........(((((.((((((((.((.....)).)))))))).)))))....)))((((....)))).)))))))).).)))))))))) ( -35.72) >DroSec_CAF1 97190 120 - 1 CGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAU .(((((((((((((((((.((((.........(....)....(((((((..(......)..))))))).)))).)))))))).(((((((....)))))......))....))))))))) ( -29.90) >DroSim_CAF1 3428 120 - 1 CGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUAUUUUUUAU .(..(..(((((((((((.((((.........(....)....(((((((..(......)..))))))).)))).))))))).))))((((....)))).....)..)............. ( -28.30) >DroEre_CAF1 19247 117 - 1 CGUGGAAAGCGCCAAC---AAAAACUUAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCACUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAU .(((((((((((((((---((......(((((............(((((..(......)..))))))))))...))))))))...(((((....)))))............))))))))) ( -28.30) >consensus CGUGGAAAGCGCCAACAAUAAAAACCCAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUGGCCGCUGCUGUUAACAGCAUUUGUUGCAGUUGUUUUUUAU .((((((((((.(..(((((((..................(((((.((((((((.((.....)).)))))))).)))))......(((((....))))))))))))..).)))))))))) (-25.61 = -26.30 + 0.69)


| Location | 21,041,970 – 21,042,090 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.52 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21041970 120 - 23771897 AGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAGCAACAAAAACCCAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCCUUUCUUGUUG .((((((((((((.((..(((.(.......))))...)).))))))))))))..((.............)).........(((((.((((((((.((.....)).)))))))).))))). ( -36.72) >DroSec_CAF1 97230 120 - 1 AGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUG .((((((((((((.((..(((.(.......))))...)).))))))))))))............................(((((.((((((((.((.....)).)))))))).))))). ( -34.00) >DroSim_CAF1 3468 120 - 1 AGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUG .((((((((((((.((..(((.(.......))))...)).))))))))))))............................(((((.((((((((.((.....)).)))))))).))))). ( -34.00) >DroEre_CAF1 19287 117 - 1 AGCGUUUUCCAUGUGAUUGCCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAAC---AAAAACUUAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUG .((((((((((((.((..(((.........)).)...)).))))))))))))....---.....................(((((.((((((((.((.....)).)))))))).))))). ( -31.60) >consensus AGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAAAAAUCAAAACAACAAAGGCGAAAAUGAAUUUGUUGCUUUUCUUGUUG .((((((((((((.((..(((.(.......))))...)).))))))))))))............................(((((.((((((((.((.....)).)))))))).))))). (-33.46 = -33.52 + 0.06)

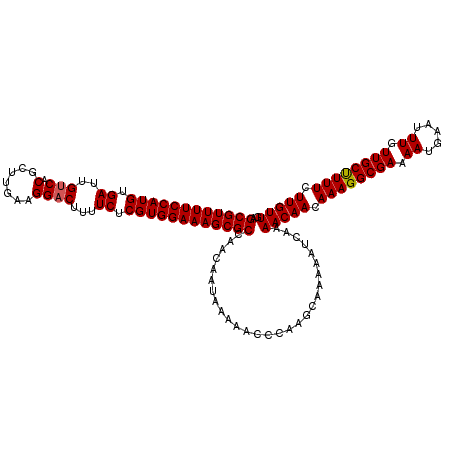
| Location | 21,042,010 – 21,042,127 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21042010 117 - 23771897 GCAAAAUGAAAACGAUAAAUCAGAAACAG---CAGCGGCGAGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAGCAACAAAAACCCAAGCAAAAAUCAA ((...........((....))........---..(..((..((((((((((((.((..(((.(.......))))...)).))))))))))))..))..)..........))......... ( -30.10) >DroSec_CAF1 97270 120 - 1 GCAAAAUGAAAACGAUAAAUCAGCAACAGCGGCAGCGGCGAGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAA ((....................((....((....)).))(.((((((((((((.((..(((.(.......))))...)).)))))))))))))................))......... ( -30.90) >DroSim_CAF1 3508 120 - 1 GCAAAAUGAAAACGAUAAAUCAGCAACAGCAGCAGCGGCGAGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAGAAAUCAA ((....................((....((....)).))(.((((((((((((.((..(((.(.......))))...)).)))))))))))))................))......... ( -30.20) >DroEre_CAF1 19327 114 - 1 GCAGAAUGAAAACGAUAAAUCAGCAGCAA---CAGCGGCGAGCGUUUUCCAUGUGAUUGCCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAAC---AAAAACUUAAGCAAAAAUCAA ((....................((.((..---..)).))(.((((((((((((.((..(((.........)).)...)).)))))))))))))...---..........))......... ( -28.50) >consensus GCAAAAUGAAAACGAUAAAUCAGCAACAG___CAGCGGCGAGCGUUUUCCAUGUGAUUGUCACGCUUGAAGGACUUUUCUCGUGGAAAGCGCCAACAAUAAAAACCCAAGCAAAAAUCAA ((...........((....))..................(.((((((((((((.((..(((.(.......))))...)).)))))))))))))................))......... (-24.40 = -24.65 + 0.25)

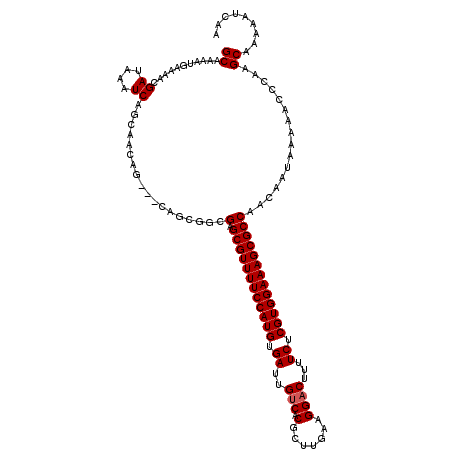
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:48 2006