| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
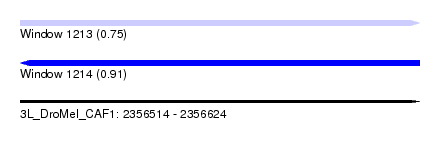
| Location | 2,356,514 – 2,356,624 |
| Length | 110 |
| Max. P | 0.908089 |

| Location | 2,356,514 – 2,356,624 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
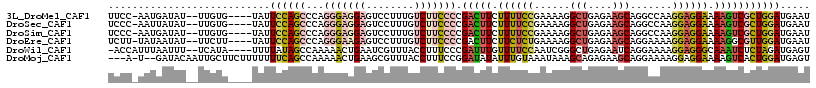
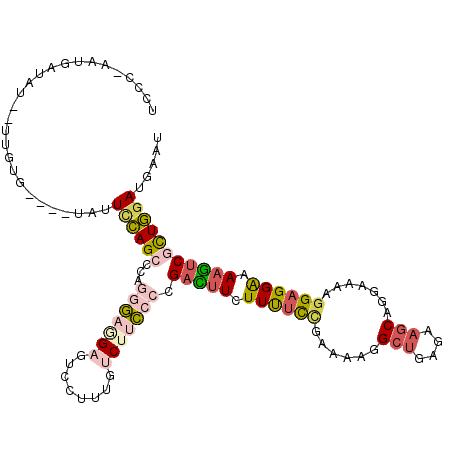
>3L_DroMel_CAF1 2356514 110 + 23771897 AUUCAUCCAGCGACUUUUCCUCCUUGGCCUGCUUCUCAGCCUUUUCGGAAAAGAAGUCGGGGAAGACAAAGGACUCCUCCCUGGGCUGGAAUA----CACAA--AUAUCAUU-GGAA .....((((((..(((((((.....(((.((.....))))).....)))))))...(((((((.((........)).)))))))))))))...----.....--........-.... ( -33.70) >DroSec_CAF1 105411 110 + 1 AUUCAUCCAGCGACUUUUCCUCCUUGGCCUGCUUCUCAGCCUUUUCGGAAAAGAAGUCGGGGAAGACAAAGGACUCCUCCCUGGGCUGGAAUA----CACAA--AUAUAAUU-GGGA .....((((((..(((((((.....(((.((.....))))).....)))))))...(((((((.((........)).)))))))))))))...----.....--........-.... ( -33.70) >DroSim_CAF1 106411 110 + 1 AUUCAUCCAGCGACUUUUCCUCCUUGGCCUGCUUCUCAGCCUUUUCGGAAAAGAAGUCGGGGAAGACAAAGGACUCCUCCCUGGGCUGGAAUA----CACAA--AUAUCAUU-GGGA .....((((((..(((((((.....(((.((.....))))).....)))))))...(((((((.((........)).)))))))))))))...----.....--........-.... ( -33.70) >DroEre_CAF1 112394 110 + 1 AUUCAUCCAACGCCUUUUCCUCCUUUUCCUGCUUCUCAGCCUUUUCAGAGAAGAAGUCGGGGAAGACAAAGGACUCUUCCCUGGGCUGGAAUA----AAGAA--AUAUUAUA-AAGA .....((((..(((..............((.((((((..........)))))).)).(((((((((........))))))))))))))))...----.....--........-.... ( -29.10) >DroWil_CAF1 198922 110 + 1 ACUCAUCUAGAGAUUUGCCCUCCUUUUCCUGAUUCUCAGCCCGAUUGGAAAACAAAUCGGGAAAGGUAAACGAUUCAGUUUUUGGCUAUAAAA----UAUGA--AAAUUAAAUGGU- .(((.....))).((((((.........(((.....)))(((((((........)))))))...))))))...((((..(((((....)))))----..)))--)...........- ( -22.00) >DroMoj_CAF1 113547 111 + 1 ACUCAUCCAGUGACUUUUCCUCCUUUUCCUGCUUCUCUGCUUUAUUUACAAAUAUAUCCGGAAAGGUAAACGCUUCAGUUUUUGGCUGAAAAAAAGAAGCAAUUGUAUC--A-U--- .......((((.((((((((..........((......))..((((....)))).....))))))))....(((((..(((((.....)))))..))))).))))....--.-.--- ( -19.00) >consensus AUUCAUCCAGCGACUUUUCCUCCUUGGCCUGCUUCUCAGCCUUUUCGGAAAAGAAGUCGGGGAAGACAAAGGACUCCUCCCUGGGCUGGAAUA____CACAA__AUAUCAUA_GGGA .....(((((((((((((..(((.......((......))......)))..)))))))(((((.((........)).)))))..))))))........................... (-16.39 = -17.29 + 0.90)

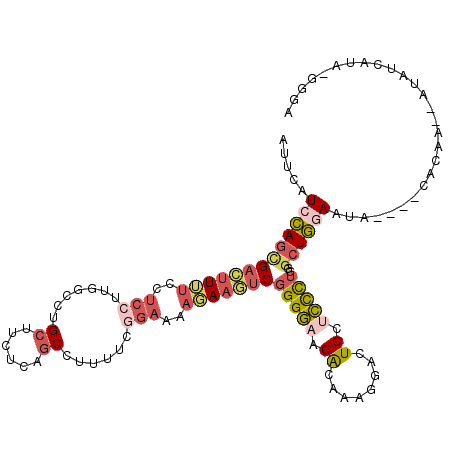
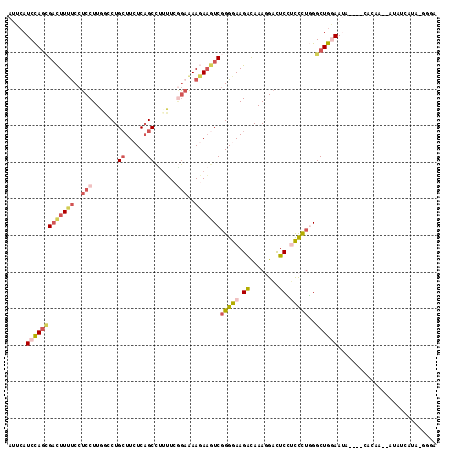
| Location | 2,356,514 – 2,356,624 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2356514 110 - 23771897 UUCC-AAUGAUAU--UUGUG----UAUUCCAGCCCAGGGAGGAGUCCUUUGUCUUCCCCGACUUCUUUUCCGAAAAGGCUGAGAAGCAGGCCAAGGAGGAAAAGUCGCUGGAUGAAU ....-........--.....----...((((((...(((((((........))))))).(((((.((((((.....((((........))))..)))))).)))))))))))..... ( -37.20) >DroSec_CAF1 105411 110 - 1 UCCC-AAUUAUAU--UUGUG----UAUUCCAGCCCAGGGAGGAGUCCUUUGUCUUCCCCGACUUCUUUUCCGAAAAGGCUGAGAAGCAGGCCAAGGAGGAAAAGUCGCUGGAUGAAU ....-........--.....----...((((((...(((((((........))))))).(((((.((((((.....((((........))))..)))))).)))))))))))..... ( -37.20) >DroSim_CAF1 106411 110 - 1 UCCC-AAUGAUAU--UUGUG----UAUUCCAGCCCAGGGAGGAGUCCUUUGUCUUCCCCGACUUCUUUUCCGAAAAGGCUGAGAAGCAGGCCAAGGAGGAAAAGUCGCUGGAUGAAU ....-........--.....----...((((((...(((((((........))))))).(((((.((((((.....((((........))))..)))))).)))))))))))..... ( -37.20) >DroEre_CAF1 112394 110 - 1 UCUU-UAUAAUAU--UUCUU----UAUUCCAGCCCAGGGAAGAGUCCUUUGUCUUCCCCGACUUCUUCUCUGAAAAGGCUGAGAAGCAGGAAAAGGAGGAAAAGGCGUUGGAUGAAU ....-........--...((----(((.(((((((.(((((((........)))))))...((((((.(((......(((....))).))).)))))).....)).)))))))))). ( -32.30) >DroWil_CAF1 198922 110 - 1 -ACCAUUUAAUUU--UCAUA----UUUUAUAGCCAAAAACUGAAUCGUUUACCUUUCCCGAUUUGUUUUCCAAUCGGGCUGAGAAUCAGGAAAAGGAGGGCAAAUCUCUAGAUGAGU -............--.....----.......(((..((((......)))).((((((((((((.(....).)))))))(((.....)))..)))))..)))....(((.....))). ( -22.80) >DroMoj_CAF1 113547 111 - 1 ---A-U--GAUACAAUUGCUUCUUUUUUUCAGCCAAAAACUGAAGCGUUUACCUUUCCGGAUAUAUUUGUAAAUAAAGCAGAGAAGCAGGAAAAGGAGGAAAAGUCACUGGAUGAGU ---.-(--(((......(((((..(((((.....)))))..))))).....(((((((.......(((((.......)))))......)).))))).......)))).......... ( -19.82) >consensus UCCC_AAUGAUAU__UUGUG____UAUUCCAGCCCAGGGAGGAGUCCUUUGUCUUCCCCGACUUCUUUUCCGAAAAGGCUGAGAAGCAGGAAAAGGAGGAAAAGUCGCUGGAUGAAU ...........................((((((...(((((((........))))))).(((((.((((((......(((....))).......)))))).)))))))))))..... (-20.74 = -21.97 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:14 2006