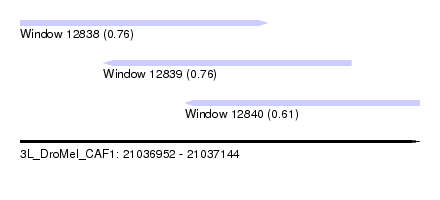
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,036,952 – 21,037,144 |
| Length | 192 |
| Max. P | 0.762808 |

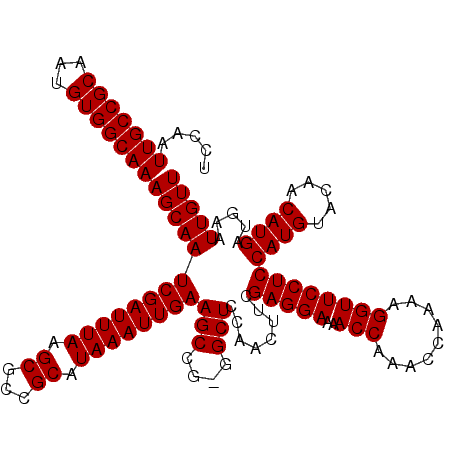
| Location | 21,036,952 – 21,037,071 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21036952 119 + 23771897 UCCAAUUGCCGCAAUGUGGCAAAGCAAUCGAUUUAAGCGCCGCAUAAAUUGAAGCCG-GGCUCCAACUUCGAGGAAAACCAAACCAAAAGGUUCCUCCAUGUACAACAUGAUGAAUUGUU (((..(((((((...))))))).((..((((((((.((...)).)))))))).)).)-))...(((.((((((((..(((.........))))))))((((.....))))..)))))).. ( -30.50) >DroSec_CAF1 92062 119 + 1 UCCAAUUGCCGCAAUGUGGCAAAGCAAUCGAUUUAAGCGCCGCAUAAAUUGAAGCCA-GGCUCCAACUUCGAGGAAAACCAAACCAAAAGGUUCCUCCAUGUACAACAUGAUGAAUUGUU .((..(((((((...))))))).((..((((((((.((...)).)))))))).))..-))...(((.((((((((..(((.........))))))))((((.....))))..)))))).. ( -29.40) >DroEre_CAF1 14099 120 + 1 UCCAAUUGCCGCAAUGUGGCAAAGCAAUCGAUUUAAGCGCCGCAUAAAUUGAAGCCGGGGCUCCAGCGCCGAGGAAAACCAAACCAAAAGGUUCCUCCAUGUACAACAUGAUGAAUUGUU .....(((((((...)))))))(((((((((((((.((...)).))))))))....((.((....)).))(((((..(((.........))))))))((((.....)))).....))))) ( -33.50) >consensus UCCAAUUGCCGCAAUGUGGCAAAGCAAUCGAUUUAAGCGCCGCAUAAAUUGAAGCCG_GGCUCCAACUUCGAGGAAAACCAAACCAAAAGGUUCCUCCAUGUACAACAUGAUGAAUUGUU .....(((((((...)))))))(((((((((((((.((...)).))))))))(((....)))........(((((..(((.........))))))))((((.....)))).....))))) (-29.70 = -29.70 + -0.00)


| Location | 21,036,992 – 21,037,111 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -39.09 |
| Energy contribution | -39.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21036992 119 - 23771897 UGCCCACUGGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUUGGUUUUCCUCGAAGUUGGAGCC-CGGCUUCAAUUUAUGCG (((((......)).)))((((((((....)))))...((((((......(((((.....)))))..(((((....))))).))))))...((((((((...)-)))))))......))). ( -38.10) >DroSec_CAF1 92102 119 - 1 UGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUUGGUUUUCCUCGAAGUUGGAGCC-UGGCUUCAAUUUAUGCG (((((......)).)))((((((((....)))))...((((((......(((((.....)))))..(((((....))))).))))))....((((((((((.-..)))))))))).))). ( -37.80) >DroEre_CAF1 14139 120 - 1 UGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUUGGUUUUCCUCGGCGCUGGAGCCCCGGCUUCAAUUUAUGCG .........(((((((....(((((....)))))...((((((......(((((.....)))))..(((((....))))).))))))....)))))(((((....))))).......)). ( -39.60) >consensus UGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUUGGUUUUCCUCGAAGUUGGAGCC_CGGCUUCAAUUUAUGCG (((((......)).)))((((((((....)))))...((((((......(((((.....)))))..(((((....))))).))))))....((((((((((....)))))))))).))). (-39.09 = -39.53 + 0.45)

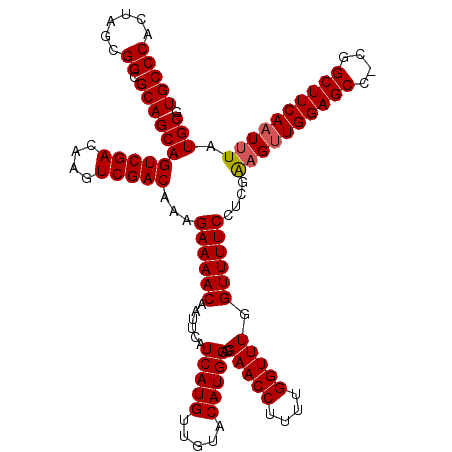
| Location | 21,037,031 – 21,037,144 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -31.47 |
| Energy contribution | -31.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21037031 113 - 23771897 GCACUUCUCUGCCACAGUUUUC-------AGUUUGCAAGUUGCCCACUGGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUU ......(((.......((((((-------....(((..((((((....))))))...)))(((((....)))))...)))))).......((((.....)))))))(((((....))))) ( -34.30) >DroSec_CAF1 92141 113 - 1 GCACUUCUCUGCCACAGUUUUC-------AGUUUGCGAGUUGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUU ......(((.......((((((-------.........(((((((......)).))))).(((((....)))))...)))))).......((((.....)))))))(((((....))))) ( -32.60) >DroEre_CAF1 14179 120 - 1 GCACUUCUCUGCCACAGUUUUCAGUUUUCAGUUUCCAAGUUGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUU ..........((((.((.((((.((((((.........(((((((......)).))))).(((((....)))))...))))))......(((((.....))))).)))).))..)))).. ( -32.80) >consensus GCACUUCUCUGCCACAGUUUUC_______AGUUUGCAAGUUGCCCACUAGCGGCGCAGCAGUCGACAAGUCGACAAAGAAAACAAUUCAUCAUGUUGUACAUGGAGGAACCUUUUGGUUU ......(((.......((((((................(((((((......)).))))).(((((....)))))...)))))).......((((.....)))))))(((((....))))) (-31.47 = -31.47 + -0.00)

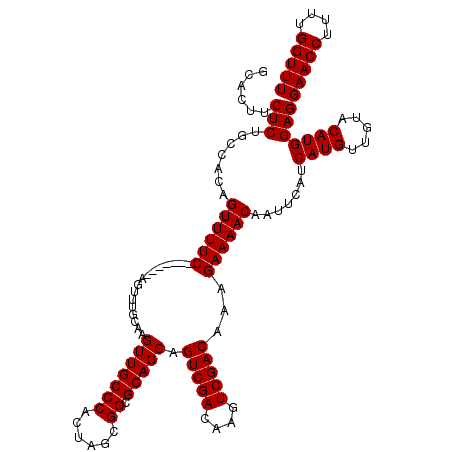
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:35 2006