
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,034,628 – 21,034,828 |
| Length | 200 |
| Max. P | 0.772800 |

| Location | 21,034,628 – 21,034,748 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21034628 120 + 23771897 GCAUUCGUUUUUGCAUUGACUCGUGUAUAUUUGGAUUUUUUUUUUUUAAAGUGCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCACCGGAUGGGCAGAUGACUCA ..(((((..(.(((((......))))).)..)))))............(((((((..(((....)))..))))))).((.((((((((((..(((......)))..))).))))))))). ( -33.90) >DroSec_CAF1 89747 115 + 1 GCAUUUGUUUUUGCAUUGAGUUGUGUAUUUUUGGAUUUU-----UUUAAUGUCCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCA (((........)))..(((((..(.........((((((-----(.....((.((..(((....)))..)).))..))))))).....((((((..(((....))))))))))..))))) ( -29.40) >DroEre_CAF1 11313 106 + 1 GCAUUUGUUUUUGCAGUGUGUUGUGUCCAUUUG--------------AAUGUGCCUUUCAAUCUUGGUUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUUGCCGGAUGGGCAGAUGACUCA (((((.((....)))))))......((((....--------------...(((((..(((....)))..))))).))))(((((((((((..(((......)))..))).)))))))).. ( -32.50) >consensus GCAUUUGUUUUUGCAUUGAGUUGUGUAUAUUUGGAUUUU_____UUUAAUGUGCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCA (((........)))....................................(((((..(((....)))..)))))...((.((((((((((..(((......)))..))).))))))))). (-27.90 = -28.23 + 0.33)


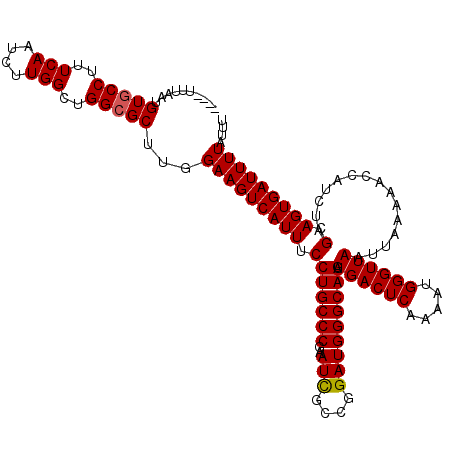
| Location | 21,034,668 – 21,034,788 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -30.25 |
| Energy contribution | -30.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21034668 120 + 23771897 UUUUUUUAAAGUGCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCACCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUU ........(((((((..(((....)))..))))))).(((((((((.(((((((..(((....))))))))).((((((.....))))))...............).))))))))).... ( -34.60) >DroSec_CAF1 89786 116 + 1 ----UUUAAUGUCCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUU ----((((((..(((......(((.(((....))).)))(((((((((((..(((......)))..))).))))))))......)))...))))))........................ ( -29.30) >DroEre_CAF1 11346 112 + 1 -------AAUGUGCCUUUCAAUCUUGGUUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUUGCCGGAUGGGCAGAUGACUCAA-AUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUU -------...(((((..(((....)))..)))))((((((((((((((((..(((......)))..))).))))))))...-...((((.......)))).))))).............. ( -32.10) >consensus ____UUUAAUGUGCCUUUCAAUCUUGGCUGGCGCUUGGAAGUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUU ..........(((((..(((....)))..)))))...(((((((((.(((((((..(((....))))))))).((((((.....))))))...............).))))))))).... (-30.25 = -30.37 + 0.11)


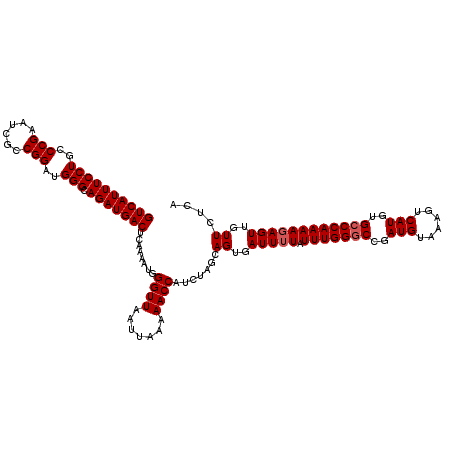
| Location | 21,034,708 – 21,034,828 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -30.97 |
| Energy contribution | -31.63 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21034708 120 + 23771897 GUCAUUUCCUGCCCGAAUCACCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUUGGGCCGAUGUAAAGUCAUGUGCCCAAAAGAGUUGUUCUCA ((((((((((..(((......)))..))).)))))))........((((.......))))....((.((..(((((.(((((((..(((......)))..))))))))))))..)))).. ( -34.90) >DroSec_CAF1 89822 120 + 1 GUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUUGGGCCGAUGUAAAGUCAUGUGCCCAAAAGAGUUGUUCUCA ((((((((((..(((......)))..))).)))))))........((((.......))))....((.((..(((((.(((((((..(((......)))..))))))))))))..)))).. ( -34.90) >DroEre_CAF1 11379 119 + 1 GUCAUUUCCUGCCCGAAUUGCCGGAUGGGCAGAUGACUCAA-AUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUUGGCCCGAUGAAAAGUCAUGUGCCCAAAACAGUUGUUCCCA ........(((.(((......))).((((((.((((((...-.((((((((.(((((..(((......))).))))).))))))))......)))))).))))))...)))......... ( -35.80) >consensus GUCAUUUCCUGCCCGAAUCGCCGGAUGGGCAGAUGACUCAAAAUGGGUUAAUUAAAAACCAUCUAGCAGUGAUUUUAUUUGGGCCGAUGUAAAGUCAUGUGCCCAAAAGAGUUGUUCUCA ((((((((((..(((......)))..))).)))))))........((((.......)))).......((..(((((.(((((((..(((......)))..))))))))))))..)).... (-30.97 = -31.63 + 0.67)


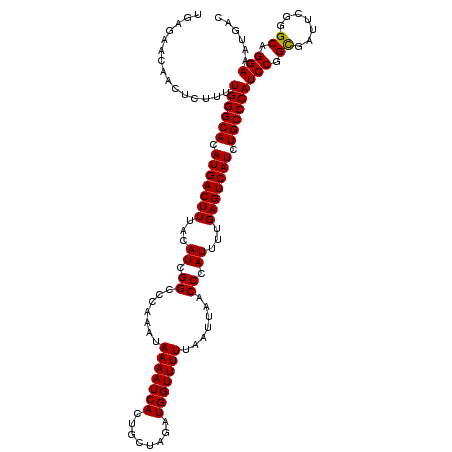
| Location | 21,034,708 – 21,034,828 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -28.96 |
| Energy contribution | -28.74 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21034708 120 - 23771897 UGAGAACAACUCUUUUGGGCACAUGACUUUACAUCGGCCCAAAUAAAAUCACUGCUAGAUGGUUUUUAAUUAACCCAUUUUGAGUCAUCUGCCCAUCCGGUGAUUCGGGCAGGAAAUGAC .(((.....))).(((((((..(((......)))..))))))).....((.(((((((((((.((......)).))))))(((((((((.........))))))))))))))))...... ( -32.60) >DroSec_CAF1 89822 120 - 1 UGAGAACAACUCUUUUGGGCACAUGACUUUACAUCGGCCCAAAUAAAAUCACUGCUAGAUGGUUUUUAAUUAACCCAUUUUGAGUCAUCUGCCCAUCCGGCGAUUCGGGCAGGAAAUGAC .(((.....))).(((((((..(((......)))..))))))).....((.((((((((((..((..(((......)))..))..)))))(((.....)))......)))))))...... ( -31.30) >DroEre_CAF1 11379 119 - 1 UGGGAACAACUGUUUUGGGCACAUGACUUUUCAUCGGGCCAAAUAAAAUCACUGCUAGAUGGUUUUUAAUUAACCCAU-UUGAGUCAUCUGCCCAUCCGGCAAUUCGGGCAGGAAAUGAC ((....)).((((..((((((.(((((((......(((......(((((((........))))))).......)))..-..))))))).)))))).((((....))))))))........ ( -33.82) >consensus UGAGAACAACUCUUUUGGGCACAUGACUUUACAUCGGCCCAAAUAAAAUCACUGCUAGAUGGUUUUUAAUUAACCCAUUUUGAGUCAUCUGCCCAUCCGGCGAUUCGGGCAGGAAAUGAC ...............((((((.(((((((...((.((.......(((((((........))))))).......)).))...))))))).))))))(((.((.......)).)))...... (-28.96 = -28.74 + -0.22)

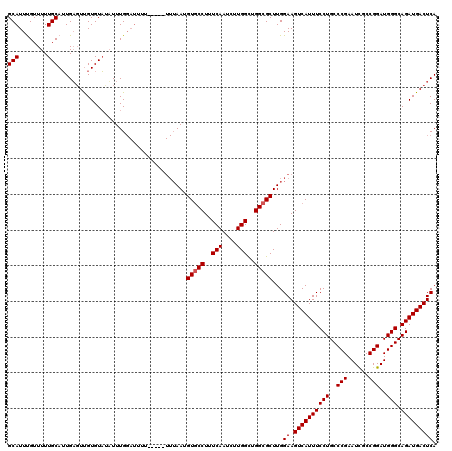
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:29 2006