
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,020,162 – 21,020,266 |
| Length | 104 |
| Max. P | 0.970444 |

| Location | 21,020,162 – 21,020,266 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.41 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21020162 104 + 23771897 UCGCCAUCAUAGUUACCGACAGCUCUCGCUCUAGCUGUCCCUCU--CUUCGCUGAUAUCUCCCAUUUAU------AUAUAUUUUCUCGGCAUCUGGGAAACUGCAUAAUUUU .........(((((.(((((((((........))))))).....--....(((((..............------..........))))).....)).)))))......... ( -19.76) >DroSim_CAF1 46222 110 + 1 UCGCCAUCAUAGUUACCGACAGCUCUCGCUCUAGCUGUCCCUCU--CUUCGCCGAUAUCUCCCAUUUAUAUGUAUAUAUAUUUUCUCGGCAUCUGGGAAACUGCAUAAUUUU .........(((((.(((((((((........))))))).....--....(((((...........(((((....))))).....))))).....)).)))))......... ( -21.79) >DroEre_CAF1 46991 102 + 1 UCGCCAUCAUAGUUACCGACAGCUCUCGCUCUAGCUGUCUCUCU--CUUUCCCGAUAUCUCCCAUUUAU--------AUAUUUUCUCGGCAUCUGGGAAACUGCAUAAUUUU ..((.............(((((((........))))))).....--.(((((((((..(..........--------..........)..))).))))))..))........ ( -19.55) >DroYak_CAF1 47199 104 + 1 UCGCCAUCAUAGUUACCGACAGCUCUCGCUCUAGCUGUCUCUCUCUCUUCACCGAUAUCUCCCAUUUAU--------AUAUUUUCUCGGCAUCUGGGAAACUGCAUAAUUUU .........(((((.(((((((((........)))))))............((((..............--------........))))......)).)))))......... ( -17.95) >consensus UCGCCAUCAUAGUUACCGACAGCUCUCGCUCUAGCUGUCCCUCU__CUUCGCCGAUAUCUCCCAUUUAU________AUAUUUUCUCGGCAUCUGGGAAACUGCAUAAUUUU .........(((((.(((((((((........)))))))...........(((((..............................))))).....)).)))))......... (-18.10 = -18.41 + 0.31)

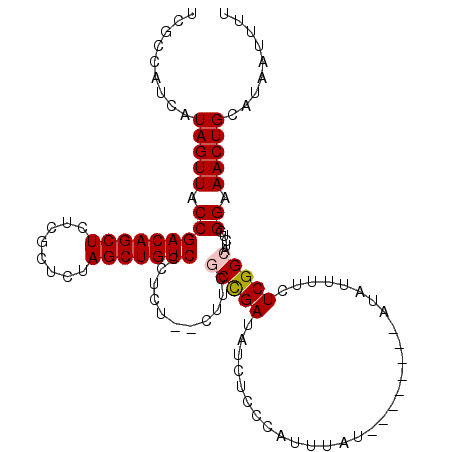
| Location | 21,020,162 – 21,020,266 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -21.95 |
| Energy contribution | -21.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21020162 104 - 23771897 AAAAUUAUGCAGUUUCCCAGAUGCCGAGAAAAUAUAU------AUAAAUGGGAGAUAUCAGCGAAG--AGAGGGACAGCUAGAGCGAGAGCUGUCGGUAACUAUGAUGGCGA .......(((.((((((((..................------.....))))))))((((....((--..(..(((((((........)))))))..)..)).)))).))). ( -22.70) >DroSim_CAF1 46222 110 - 1 AAAAUUAUGCAGUUUCCCAGAUGCCGAGAAAAUAUAUAUACAUAUAAAUGGGAGAUAUCGGCGAAG--AGAGGGACAGCUAGAGCGAGAGCUGUCGGUAACUAUGAUGGCGA ...(((((..((((.((....((((((.....(((.(((....))).))).......))))))...--.....(((((((........))))))))).)))))))))..... ( -24.40) >DroEre_CAF1 46991 102 - 1 AAAAUUAUGCAGUUUCCCAGAUGCCGAGAAAAUAU--------AUAAAUGGGAGAUAUCGGGAAAG--AGAGAGACAGCUAGAGCGAGAGCUGUCGGUAACUAUGAUGGCGA .......(((..((((((.((((((.(........--------.....).))...)))))))))).--.....(((((((........))))))).............))). ( -23.02) >DroYak_CAF1 47199 104 - 1 AAAAUUAUGCAGUUUCCCAGAUGCCGAGAAAAUAU--------AUAAAUGGGAGAUAUCGGUGAAGAGAGAGAGACAGCUAGAGCGAGAGCUGUCGGUAACUAUGAUGGCGA .......(((.((((((((................--------.....))))))))((((......((..(..(((((((........)))))))..)..)).)))).))). ( -22.60) >consensus AAAAUUAUGCAGUUUCCCAGAUGCCGAGAAAAUAU________AUAAAUGGGAGAUAUCGGCGAAG__AGAGAGACAGCUAGAGCGAGAGCUGUCGGUAACUAUGAUGGCGA .......(((.((((((((.............................)))))))).................(((((((........))))))).............))). (-21.95 = -21.95 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:08 2006