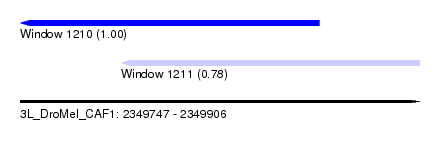
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,349,747 – 2,349,906 |
| Length | 159 |
| Max. P | 0.995831 |

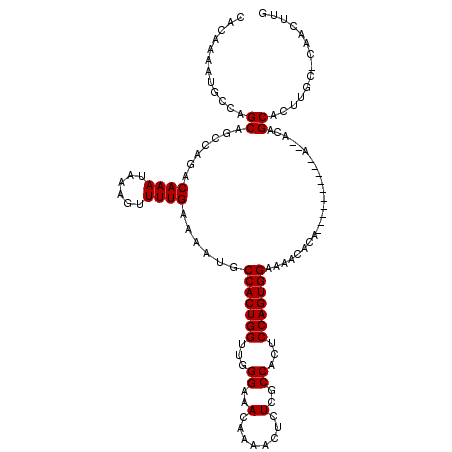
| Location | 2,349,747 – 2,349,866 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2349747 119 - 23771897 GCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAACACACGCAUAUCACACAACAGCACUUGCCCAACUUGCUUUGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .(((((((.(((((..........)).)))..)))))))........................(((...((.......))..)))........(((((((((......))))))))).. ( -21.70) >DroSec_CAF1 98735 102 - 1 GCCACUGGUUGGGUAACAAAACUCUCGCCACUCCAGUGGAAAACACA--------------CAGCACUUGC-CAACUUGU--UGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .....((((.(((((((((........((((....))))........--------------..((....))-....))))--)))))))))..(((((((((......))))))))).. ( -22.60) >DroSim_CAF1 99735 102 - 1 GCCACUGGUUGGGUAACAAAACUCUCGCCACUCCAGUGGAAAACACA--------------CAGCACUUGC-CAACUUGC--UGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .(((((((...(((............)))...)))))))........--------------(((((.....-.....)))--)).........(((((((((......))))))))).. ( -23.50) >DroEre_CAF1 105553 104 - 1 GCCACUGGUUGGGAAACAAAAUUGUCGCCACUCCAGUGGAAAACAC-----------AACACAGCACUUGC-CAACU-GC--UGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .(((((((...((..(((....)))..))...))))))).......-----------....(((((.....-....)-))--)).........(((((((((......))))))))).. ( -26.20) >DroYak_CAF1 102566 108 - 1 GCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAAUACACA-------AAACACAGCACUUGC-CAACU-GC--UGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .(((((((.(((((..........)).)))..)))))))..........-------.....(((((.....-....)-))--)).........(((((((((......))))))))).. ( -23.60) >consensus GCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAACACA__________A__ACAGCACUUGC_CAACUUGC__UGCCUAUCAAUUUAUUUAGAAUUUCAUUUGAAUAAUU .(((((((...((..(........)..))...)))))))........................(((...((.......))..)))........(((((((((......))))))))).. (-20.22 = -20.06 + -0.16)

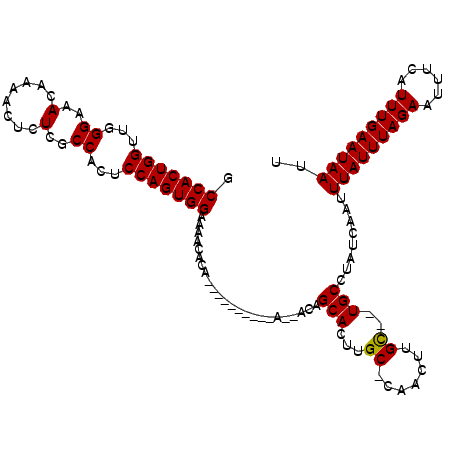
| Location | 2,349,787 – 2,349,906 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -23.29 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2349787 119 - 23771897 CACAAAAUGCCAGCAGCCAGACAAAUAAAGUUUUGAAAAUGCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAACACACGCAUAUCACACAACAGCACUUGCCCAACUUG ......((((...........((((......))))......(((((((.(((((..........)).)))..))))))).........))))...........((....))........ ( -21.00) >DroSec_CAF1 98773 104 - 1 CACAAAAUGCCAGCAGGCAGACAAAUAAAGUUUUGAAAAUGCCACUGGUUGGGUAACAAAACUCUCGCCACUCCAGUGGAAAACACA--------------CAGCACUUGC-CAACUUG ...............(((((.((((......))))......(((((((...(((............)))...)))))))........--------------......))))-)...... ( -23.90) >DroSim_CAF1 99773 104 - 1 CACAAAAUGCCAGCAGGCAGACAAAUAAAGUUUUGAAAAUGCCACUGGUUGGGUAACAAAACUCUCGCCACUCCAGUGGAAAACACA--------------CAGCACUUGC-CAACUUG ...............(((((.((((......))))......(((((((...(((............)))...)))))))........--------------......))))-)...... ( -23.90) >DroEre_CAF1 105591 106 - 1 CACAAAAUGCCAGCAGCCAGACAAAUAAAGUUUUGAAAGUGCCACUGGUUGGGAAACAAAAUUGUCGCCACUCCAGUGGAAAACAC-----------AACACAGCACUUGC-CAACU-G ..........(((..((.((.((((......))))...((.(((((((...((..(((....)))..))...)))))))...))..-----------.........)).))-...))-) ( -24.60) >DroYak_CAF1 102604 110 - 1 CACAAAAUGCCAGCAGCCAGACAAAUAAUGUUUUGAAAAUGCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAAUACACA-------AAACACAGCACUUGC-CAACU-G .................(((........(((((((......(((((((.(((((..........)).)))..)))))))........))-------)))))..((....))-...))-) ( -23.04) >consensus CACAAAAUGCCAGCAGCCAGACAAAUAAAGUUUUGAAAAUGCCACUGGUUGGGAAACAAAACUCUCGCCACUCCAGUGGAAAACACA__________A__ACAGCACUUGC_CAACUUG ............((.......((((......))))......(((((((...((..(........)..))...)))))))........................)).............. (-18.58 = -18.58 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:11 2006