| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,016,314 – 21,016,461 |
| Length | 147 |
| Max. P | 0.961344 |

| Location | 21,016,314 – 21,016,421 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.27 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -11.57 |
| Energy contribution | -12.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
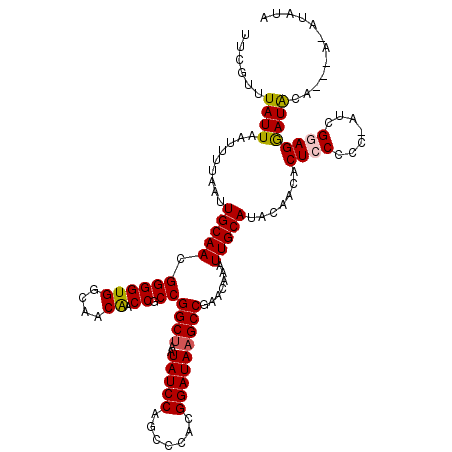
>3L_DroMel_CAF1 21016314 107 - 23771897 GCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC-AUCGGAGGAUACAAACACAUACAUACAAUGUAUACAUAGU------------ACGAAUAUCGA ..(((((..(((((.......))))))))))...........(((....((((...-...))))...........((((((...)))))).....))------------)((.....)). ( -21.30) >DroSim_CAF1 42215 95 - 1 GCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC-AUCGCAGGAUACAAACAUAUAUA-------CAUACAUAGU------------AUGAAU----- ..(((((..(((((.......)))))))))).........(((((.........((-......))...........(((.-------.....)))))------------)))...----- ( -16.50) >DroYak_CAF1 43233 89 - 1 GCCGGCUAAUAUCCAGCCCACGGAUAAGCCUAACAAAUUGCAUAUAACACUCCCCC-AUCGGAGAAUAUA------UAUA-------C-----UGGU------------ACGAAUAUCGA (((((((..(((((.......)))))))))...........(((((...((((...-...))))..))))------)...-------.-----.)))------------.((.....)). ( -19.40) >DroAna_CAF1 30308 105 - 1 CCCGGC--GUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAAACAUCACUGCCCCUCUUGGAGGAUGGAGG-AUAGAGG-------C-----UGGAGGAUAGAGGAUGUCGGAUGUCGG .(((((--((.((((((((.(((.....)))........(((........))).(((((........)))))-...).))-------)-----)))).((((.....))))..))))))) ( -35.10) >consensus GCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC_AUCGGAGGAUACAAA_AUAUAUA_______C_____UAGU____________ACGAAUAUCGA ..(((((..(((((.......))))))))))..................((((.......))))........................................................ (-11.57 = -12.58 + 1.00)

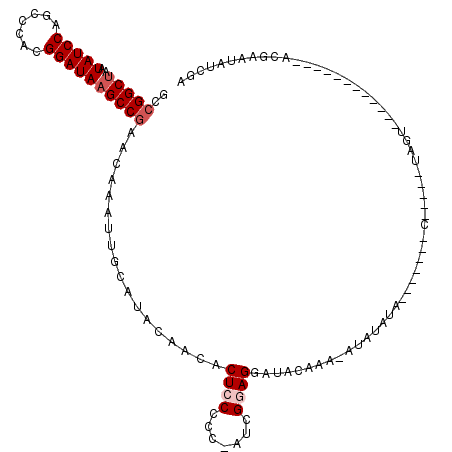
| Location | 21,016,342 – 21,016,461 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.84 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21016342 119 + 23771897 UGUAUGUGUUUGUAUCCUCCGAU-GGGGGAGUGUUGUAUGCAAUUUGUUCGGCUUAUCCGUGGGCUGGAUAUUAGCCGGCGGUUGUUGCCACCCCGUUGCAAUUAAAAUUAAUAAACGAA ......(((((((......((((-(((((((.((((....))))...)))(((....((((.(((((.....))))).)))).....))).)))))))).(((....))).))))))).. ( -37.60) >DroSim_CAF1 42231 119 + 1 UAUAUAUGUUUGUAUCCUGCGAU-GGGGGAGUGUUGUAUGCAAUUUGUUCGGCUUAUCCGUGGGCUGGAUAUUAGCCGGCGGUUGUUGCCACCCCGUUGCAAUUAAAAUUAAUAAACGAA ......(((((((....((((((-(((((((.((((....))))...)))(((....((((.(((((.....))))).)))).....))).))))))))))..((....))))))))).. ( -42.20) >DroEre_CAF1 42811 109 + 1 ----------UAUAUCCUCCGAU-GGGGGAGUGUUGUGUGCAAUUUGUUCGGCUUAUCCGUGGGCUGGAUAUUAGCCGGCGGUUGUUGCCACCCCGUUGCAAUUAAAAUUAAUAAACGAA ----------.........((((-(((((((.((((....))))...)))(((....((((.(((((.....))))).)))).....))).))))))))..................... ( -36.20) >DroYak_CAF1 43249 113 + 1 UAUA------UAUAUUCUCCGAU-GGGGGAGUGUUAUAUGCAAUUUGUUAGGCUUAUCCGUGGGCUGGAUAUUAGCCGGCGGUUGUUGCCACCCCGUUGCAAUUAAAAUUAAUAAACGAA ((((------.(((((((((...-.)))))))))))))((((((......(((....((((.(((((.....))))).)))).....))).....))))))................... ( -35.90) >DroAna_CAF1 30336 117 + 1 CCUCUAU-CCUCCAUCCUCCAAGAGGGGCAGUGAUGUUUGCAAUUUGUUCGGCUUAUCCGUGGGCUGGAUAC--GCCGGGGGGCGUUAACACCCCGUUGCAAUUAAAAUUAAUAAACGAA ....(((-(((((..((((...)))))).)).)))).(((((((....(((((.((((((.....)))))).--)))))((((.(....).))))))))))).................. ( -36.40) >consensus UAUAU_U___UAUAUCCUCCGAU_GGGGGAGUGUUGUAUGCAAUUUGUUCGGCUUAUCCGUGGGCUGGAUAUUAGCCGGCGGUUGUUGCCACCCCGUUGCAAUUAAAAUUAAUAAACGAA ..............((((((....))))))........((((((......(((....((((.((((((...)))))).)))).....))).....))))))................... (-26.72 = -27.84 + 1.12)


| Location | 21,016,342 – 21,016,461 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21016342 119 - 23771897 UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGGCAACAACCGCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC-AUCGGAGGAUACAAACACAUACA ...((((............((((((((..((....)).))).(((((..(((((.......))))))))))......))))).......((((...-...)))).....))))....... ( -30.80) >DroSim_CAF1 42231 119 - 1 UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGGCAACAACCGCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC-AUCGCAGGAUACAAACAUAUAUA ...((((............((((((((..((....)).))).(((((..(((((.......))))))))))......)))))............((-......))....))))....... ( -26.50) >DroEre_CAF1 42811 109 - 1 UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGGCAACAACCGCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCACACAACACUCCCCC-AUCGGAGGAUAUA---------- ...................((((((((..((....)).))).(((((..(((((.......))))))))))......))))).......((((...-...))))......---------- ( -28.80) >DroYak_CAF1 43249 113 - 1 UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGGCAACAACCGCCGGCUAAUAUCCAGCCCACGGAUAAGCCUAACAAAUUGCAUAUAACACUCCCCC-AUCGGAGAAUAUA------UAUA ...................(((((...(((((......)))))((((..(((((.......))))))))).......))))).......((((...-...))))......------.... ( -27.50) >DroAna_CAF1 30336 117 - 1 UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGUUAACGCCCCCCGGC--GUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAAACAUCACUGCCCCUCUUGGAGGAUGGAGG-AUAGAGG ..(.((((((........((((((.((((((....)))))).((((--.(((((.......))))).))))......)))))).((((.((.((......))))))))...)-))))).) ( -39.00) >consensus UUCGUUUAUUAAUUUUAAUUGCAACGGGGUGGCAACAACCGCCGGCUAAUAUCCAGCCCACGGAUAAGCCGAACAAAUUGCAUACAACACUCCCCC_AUCGGAGGAUACA___A_AUAUA ......((((.........(((((.((((((....)).)).))((((..(((((.......))))))))).......))))).......((((.......))))))))............ (-23.92 = -24.04 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:05 2006