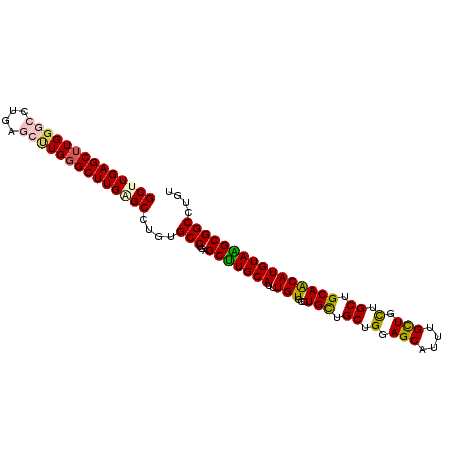
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
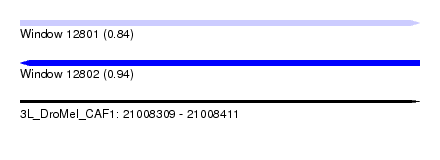
| Location | 21,008,309 – 21,008,411 |
| Length | 102 |
| Max. P | 0.938015 |

| Location | 21,008,309 – 21,008,411 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.05 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21008309 102 + 23771897 ACAGGCCGCCUACAUGUUGCAGCAACAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCUCAGGCUCAGGCUCAAGCCCAAGCUCAGGCCCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))..)))).........((((..((((........))))..))))..))))........... ( -34.40) >DroSec_CAF1 25941 102 + 1 ACAGGCCGCUUACAUGUUGCAGCAACAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCUCAGGCACAGGCUCAAGCCCAAGCUCAGGCCCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))(((........)))))))...(((...(((....)))...)))..))))........... ( -30.30) >DroSim_CAF1 34460 102 + 1 ACAGGCCGCUUACAUGUUGCAGCAACAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCCCAGGCACAGGCUCAAGCCCAAGCUCAGGCCCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))..))))......((..((....))...(((....)))...))...))))........... ( -30.60) >DroEre_CAF1 34500 96 + 1 GCAGGCCGCUUACAUGCUGCAGCAGCAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCUCAGGCCCAAGCUCAAGCCCAA------GCCCAAGCUCAGGC ((.((((((((...((((((.(.((((.....)))).).)))))).(((....)))))))...))))...))....(((..(------((....)))..))) ( -33.60) >DroYak_CAF1 31812 96 + 1 ACAGGCCGCUUACAUGUUGCAGCAGCAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCUCAGGCACAGGCCCAAGCACAG------GCCCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))..))))..........(((..(((....)))..)))...)------)))........... ( -31.50) >DroAna_CAF1 22185 102 + 1 UCAGGCCGCCUACAUGCUGCAGCAGCAACAGAUGCUCCAGCAACAGGCACAAUUGCAGGCUCAGGCGCAGGCACAAGCCCAGGCCCAGGCUCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))..)))).........((((...((....))...))))..))))..(((....)))..... ( -36.20) >consensus ACAGGCCGCUUACAUGUUGCAGCAACAGCAAAUGCUCCAGCAGCAGGCACAAAUGCAAGCUCAGGCACAGGCUCAAGCCCAAGCUCAGGCCCAAGCUCAAGC ...((((((((...(((((.((((........)))).)))))..))))..........(((..(((....)))..))).........))))........... (-27.08 = -27.05 + -0.03)

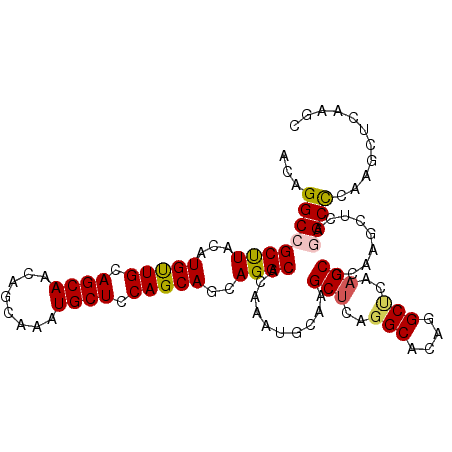
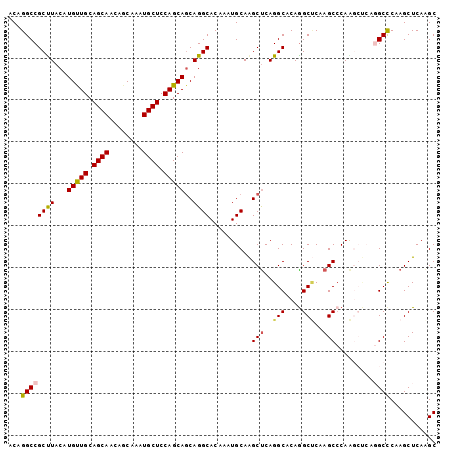
| Location | 21,008,309 – 21,008,411 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -36.74 |
| Energy contribution | -36.77 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21008309 102 - 23771897 GCUUGAGCUUGGGCCUGAGCUUGGGCUUGAGCCUGAGCCUGAGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGUUGCUGCAACAUGUAGGCGGCCUGU ((....))..(((((.(((((..(((((......)))))..)))))........((((((((.((.((((........)))).)).)).))))))))))).. ( -47.80) >DroSec_CAF1 25941 102 - 1 GCUUGAGCUUGGGCCUGAGCUUGGGCUUGAGCCUGUGCCUGAGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGUUGCUGCAACAUGUAAGCGGCCUGU (((..((((..(((....)))..))))..)))....(((...(((((((..(((...(((.((.(.(((....))).).)).)))))))))))))))).... ( -42.70) >DroSim_CAF1 34460 102 - 1 GCUUGAGCUUGGGCCUGAGCUUGGGCUUGAGCCUGUGCCUGGGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGUUGCUGCAACAUGUAAGCGGCCUGU (((..((((..(((....)))..))))..)))....(((...(((((((..(((...(((.((.(.(((....))).).)).)))))))))))))))).... ( -42.70) >DroEre_CAF1 34500 96 - 1 GCCUGAGCUUGGGC------UUGGGCUUGAGCUUGGGCCUGAGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGCUGCUGCAGCAUGUAAGCGGCCUGC (((.(((((..(((------(..(((....)))..))))..)))))((.....(((.((((((.(.((((.....)))).).)))))).))).))))).... ( -50.40) >DroYak_CAF1 31812 96 - 1 GCUUGAGCUUGGGC------CUGUGCUUGGGCCUGUGCCUGAGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGCUGCUGCAACAUGUAAGCGGCCUGU ((((.(((..((((------(((((((..(((....)))..)))..)))...).))))...))).))))....((((((((........)).)))))).... ( -38.20) >DroAna_CAF1 22185 102 - 1 GCUUGAGCUUGAGCCUGGGCCUGGGCUUGUGCCUGCGCCUGAGCCUGCAAUUGUGCCUGUUGCUGGAGCAUCUGUUGCUGCUGCAGCAUGUAGGCGGCCUGA ((((......))))..(((((.(((((((((....)))..))))))........((((..(((((.((((.(....).)))).)))))...))))))))).. ( -43.40) >consensus GCUUGAGCUUGGGCCUGAGCUUGGGCUUGAGCCUGUGCCUGAGCUUGCAUUUGUGCCUGCUGCUGGAGCAUUUGCUGCUGCUGCAACAUGUAAGCGGCCUGU ((((((((((((((....))))))))))))))....(((...(((((((..(((...(((.((.(.(((....))).).)).)))))))))))))))).... (-36.74 = -36.77 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:58 2006