| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,006,385 – 21,006,492 |
| Length | 107 |
| Max. P | 0.851408 |

| Location | 21,006,385 – 21,006,492 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -48.19 |
| Consensus MFE | -28.63 |
| Energy contribution | -30.97 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
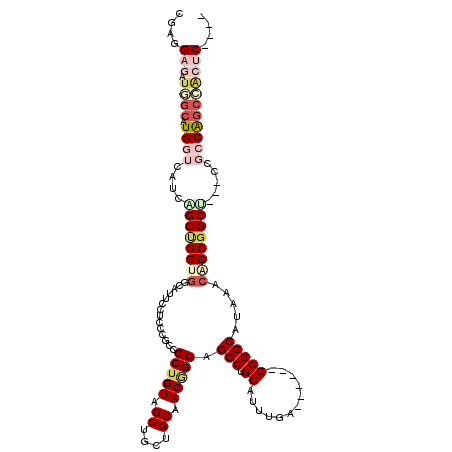
>3L_DroMel_CAF1 21006385 107 - 23771897 CGAGGAGAUAGCAUGGUCAUCCGCUGGUGGCAUUCCUCCUGCCGCUGUAGCUGCUGCAACGGCAGCCUGCAUUUGA------GCGGCAUAAACGCCGGCU---CCUCCAGCCACUC---- .((((((...((...((((((....))))))........(((((((((((((((((...)))))).)))).....)------)))))).....))...))---)))).........---- ( -42.20) >DroSec_CAF1 24018 107 - 1 CGAGGAGAUGGCAUGGUCAUCAGCUGGUGGCAUUCCUCCUGCCGCUGUAGCUGCUGCAACGGCAGCCUGCAUUUGA------GCGGCAUAAACACCUGCU---CCGCCAGCCACUC---- .(((..((((((...)))))).((((((((((.......(((((((((((((((((...)))))).)))).....)------))))))........))).---.)))))))..)))---- ( -44.26) >DroSim_CAF1 32537 107 - 1 CGAGGAGAUGGCAUGGUCAUCAGCUGGUGGCAUUCCUCCUGCCGCUGUAGCCGCUGCAACGGCAGCCUGCAUUUGA------GCGGCAUAAACACCGGCU---CCGGCAGCCACUC---- ....(((.((((...(((...((((((((..........(((((.(((((...))))).)))))(((.((......------))))).....))))))))---..))).)))))))---- ( -44.10) >DroEre_CAF1 32603 107 - 1 CGAGGAGAUGGCAUGGUCGUCUGCUGGUGGUAUUCCUCCCGCCGCUGUGGCUGCUGCAACGGCAGCCUGCAUUUGA------GCGGCAUACACACCGGCU---CCUCCAGCCACUC---- ....(((.((((.(((..(...(((((((((((.......((((((((((((((((...)))))))).)).....)------))))))))).))))))).---)..))))))))))---- ( -50.01) >DroWil_CAF1 2727 120 - 1 CGAGGAUAUUGCAUGAUCAUCCGCCGGCGGUAUACCGCCCACAGCUGUGGCAGCUGCCACAGCAGCCUGCAUCUGGGCAGCAGCGGCAUACUGACCGGCGACAACGCCAGCGGCGCUAAC ...(((((((....))).))))(((((((((((.((((.....((((((((....)))))))).((((......))))....)))).)))))).))))).....((((...))))..... ( -57.20) >DroAna_CAF1 20363 101 - 1 CGAGGAUAUGGCAUGGUCGUCGGCCGGCGGCAGGCCACCCACGGCCGUGGCAGCUGCCACUGCGGCCUGCAUCUGG------GCGGCAUAGCCACCGGCU---CCGCCG------C---- (((.(((........))).)))(((((.(((.((((......))))(((((....)))))(((.((((......))------)).)))..))).))))).---......------.---- ( -51.40) >consensus CGAGGAGAUGGCAUGGUCAUCAGCUGGUGGCAUUCCUCCCGCCGCUGUAGCUGCUGCAACGGCAGCCUGCAUUUGA______GCGGCAUAAACACCGGCU___CCGCCAGCCACUC____ ....(((.((((.((((....((((((((..............(((((.((....)).))))).(((.((............))))).....)))))))).....))))))))))).... (-28.63 = -30.97 + 2.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:53 2006