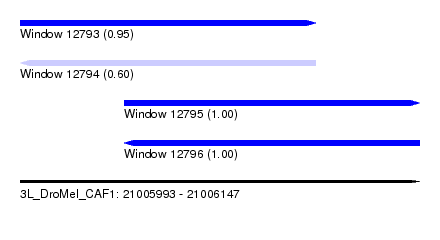
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 21,005,993 – 21,006,147 |
| Length | 154 |
| Max. P | 0.999475 |

| Location | 21,005,993 – 21,006,107 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -26.61 |
| Energy contribution | -27.90 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
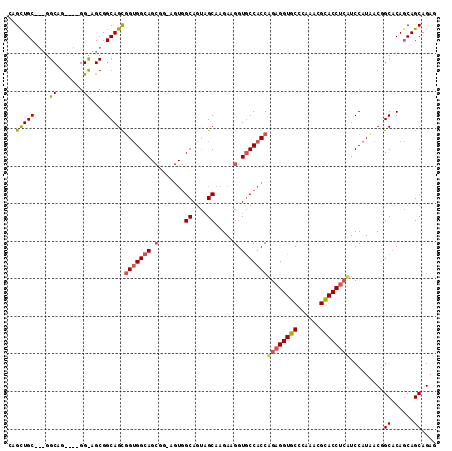
>3L_DroMel_CAF1 21005993 114 + 23771897 CAGUUGCAGUGGCAGCACAGGUAGCGGCAGCGGAGGCAGCGGGAGUGGCAGUAGCAAGAAGGUGCCACCAGAGGUGCCCAAACGCACCAGUUCCAUUACGGCACAACAGCAGAC ..((((.(((((.(((...(((.(((......(.((((.(..(.((((((.(........).)))))))...).)))))...)))))).)))))))).))))............ ( -36.60) >DroGri_CAF1 18376 102 + 1 CAGCUGC---GGCAGUCAGGGCAGCGGCAGUGGUGGCA---------GCAGCAGCAAGAAGGUACCACCGGAGGUGCCCAAGCGCACCUCCUCCAUCACCGCACAGCAGCAGAG ..(((((---.((.((....)).)).((.(((((((..---------((....)).....((.....))((((((((......)))))))).))))))).))...))))).... ( -44.10) >DroWil_CAF1 2354 96 + 1 GAGCUGC------------------GGCAGCAGUGGCAGCGGCAGUGGCAGUGGCAAGAAGGUGCCACCAGAGGUACCCAAACGUACCUCAUCCAUAACGGCCCAGCAGCAGAG ..(((((------------------.((.((....)).))(((.((((..((((((......))))))..(((((((......)))))))..))))....)))..))))).... ( -40.90) >consensus CAGCUGC___GGCAG____GG_AGCGGCAGCGGUGGCAGCGG_AGUGGCAGUAGCAAGAAGGUGCCACCAGAGGUGCCCAAACGCACCUCAUCCAUAACGGCACAGCAGCAGAG ..(((((....((.......))....)))))(((((((.(.......((....)).....).)))))))((((((((......)))))))).........((......)).... (-26.61 = -27.90 + 1.29)

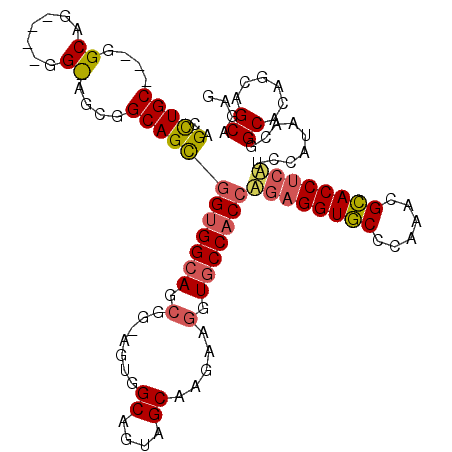
| Location | 21,005,993 – 21,006,107 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21005993 114 - 23771897 GUCUGCUGUUGUGCCGUAAUGGAACUGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCUACUGCCACUCCCGCUGCCUCCGCUGCCGCUACCUGUGCUGCCACUGCAACUG ((.(((.((.(.((.(((..((...(((((((...(((((.(...(((((((............)))))))...).))))).))).))))...))..))).))))).))))).. ( -33.90) >DroGri_CAF1 18376 102 - 1 CUCUGCUGCUGUGCGGUGAUGGAGGAGGUGCGCUUGGGCACCUCCGGUGGUACCUUCUUGCUGCUGC---------UGCCACCACUGCCGCUGCCCUGACUGCC---GCAGCUG ....(((((...(((((..(((.((((((((......))))))))((..(((.(........).)))---------..)).)))..))))).((.......)).---))))).. ( -47.80) >DroWil_CAF1 2354 96 - 1 CUCUGCUGCUGGGCCGUUAUGGAUGAGGUACGUUUGGGUACCUCUGGUGGCACCUUCUUGCCACUGCCACUGCCGCUGCCACUGCUGCC------------------GCAGCUC ....(((((..(((.((..(((..(((((((......))))))).(((((((......))))))).............)))..)).)))------------------))))).. ( -39.90) >consensus CUCUGCUGCUGUGCCGUAAUGGAAGAGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCUACUGCCACU_CCGCUGCCACCGCUGCCGCU_CC____CUGCC___GCAGCUG ....(((((...((.((..(((.((((((((......))))))))(((((((......))))))).............)))..)).)).((..........))....))))).. (-28.20 = -28.27 + 0.07)



| Location | 21,006,033 – 21,006,147 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -27.57 |
| Energy contribution | -29.47 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21006033 114 + 23771897 GGGAGUGGCAGUAGCAAGAAGGUGCCACCAGAGGUGCCCAAACGCACCAGUUCCAUUACGGCACAACAGCAGACACAGCUUCUUUUGCUGCAGCGCCAGACACCGCCGCCUCCU ((..(((((.(((((((((..(((((......(((((......))))).((......)))))))...(((.......)))..))))))))).((....).)...)))))..)). ( -42.70) >DroGri_CAF1 18411 107 + 1 -------GCAGCAGCAAGAAGGUACCACCGGAGGUGCCCAAGCGCACCUCCUCCAUCACCGCACAGCAGCAGAGCCAGCUGCUGAUGAUGCAGCGACAGACGCCGCCGCCACCC -------((.((.(((....(((......((((((((......))))))))......))).(((((((((.......))))))).)).))).(((.....))).)).))..... ( -41.20) >DroWil_CAF1 2376 114 + 1 GGCAGUGGCAGUGGCAAGAAGGUGCCACCAGAGGUACCCAAACGUACCUCAUCCAUAACGGCCCAGCAGCAGAGUCAAUUGCUUCUCUUGCAACGACAGACACCGCCGCCGCCA (((.(((((.(((((((((.((.(((....(((((((......))))))).........))))).(.(((((......))))).)))))))..(....).))).))))).))). ( -44.32) >consensus GG_AGUGGCAGUAGCAAGAAGGUGCCACCAGAGGUGCCCAAACGCACCUCAUCCAUAACGGCACAGCAGCAGAGACAGCUGCUUAUGAUGCAGCGACAGACACCGCCGCCACCA (((.(((((.((.((.....((.....))((((((((......)))))))).................)).......(((((.......))))).......)).))))).))). (-27.57 = -29.47 + 1.90)

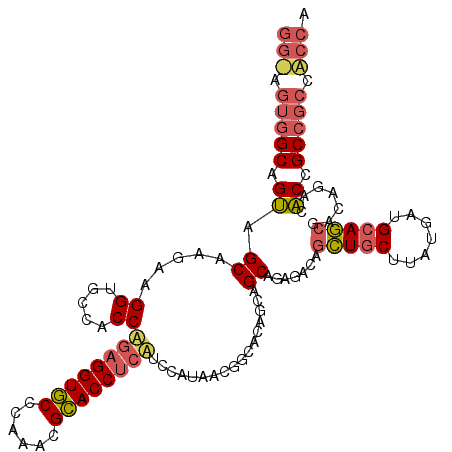

| Location | 21,006,033 – 21,006,147 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Mean single sequence MFE | -53.65 |
| Consensus MFE | -39.03 |
| Energy contribution | -39.27 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 21006033 114 - 23771897 AGGAGGCGGCGGUGUCUGGCGCUGCAGCAAAAGAAGCUGUGUCUGCUGUUGUGCCGUAAUGGAACUGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCUACUGCCACUCCC .((((..(((((((((((((((.((((((..............)))))).)))))......((((......)))))))))))...(((((((......)))))))))).)))). ( -49.14) >DroGri_CAF1 18411 107 - 1 GGGUGGCGGCGGCGUCUGUCGCUGCAUCAUCAGCAGCUGGCUCUGCUGCUGUGCGGUGAUGGAGGAGGUGCGCUUGGGCACCUCCGGUGGUACCUUCUUGCUGCUGC------- .....(((((((((((((((((((((....(((((((.......)))))))))))))))))))((((((((......))))))))((.....))....)))))))))------- ( -60.50) >DroWil_CAF1 2376 114 - 1 UGGCGGCGGCGGUGUCUGUCGUUGCAAGAGAAGCAAUUGACUCUGCUGCUGGGCCGUUAUGGAUGAGGUACGUUUGGGUACCUCUGGUGGCACCUUCUUGCCACUGCCACUGCC .(((((((((((.(((....(((((.......))))).))).))))))))..)))((..(((..(((((((......))))))).(((((((......))))))).)))..)). ( -51.30) >consensus AGGAGGCGGCGGUGUCUGUCGCUGCAACAAAAGCAGCUGACUCUGCUGCUGUGCCGUAAUGGAAGAGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCUACUGCCACU_CC .(((((((((((.(((....(((((.......))))).))).))))))))..)))........((((((((......))))))))(((((((......)))))))......... (-39.03 = -39.27 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:52 2006