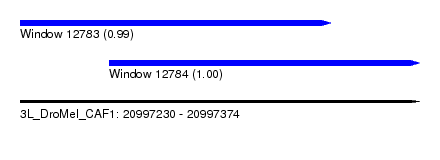
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,997,230 – 20,997,374 |
| Length | 144 |
| Max. P | 0.999003 |

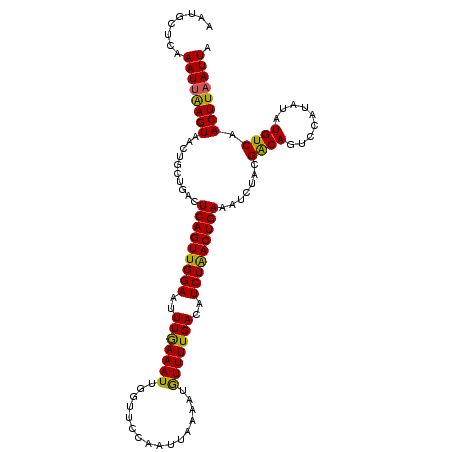
| Location | 20,997,230 – 20,997,342 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -22.43 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20997230 112 + 23771897 --------UAGCGUGGUUAUUUAAUGAUUGCGAUCAACGAAAUUCUUAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA --------....(..((((((((((((((.((.....)).)))).....))))))))))..)....(((((((((......(((((....)))))....(((....)))))))))))).. ( -25.60) >DroSec_CAF1 14802 112 + 1 --------UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUCGUUCCAAUUAAAAUAUUUUGACAUCUAACUGAAA --------....(..((((((((((....(((.((....)).)))....))))))))))..)....(((((((((..(..((((...((......))...))))..)..))))))))).. ( -26.50) >DroSim_CAF1 22245 112 + 1 --------UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA --------....(..((((((((((....(((.((....)).)))....))))))))))..)....(((((((((......(((((....)))))....(((....)))))))))))).. ( -28.30) >DroEre_CAF1 23551 112 + 1 --------UAGCGUGGUUAUUUAAUGAUUGAGUUUAACGAAAUGCUAAAAUGGAGUAGCUGCUGACUCAGUUGGAAUUUGAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUGACUGAAA --------....(..(((((((.........((((....)))).........)))))))..)....(((((..((..(..((((................))))..)..))..))))).. ( -21.36) >DroYak_CAF1 21000 120 + 1 UCUGCAUCUAGCGUGGUUAUUUAAUGAUUGAGUUAAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUAAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA ...((.....))(..((((((((((..((((((..........))))))))))))))))..)....(((((((((..(((((((................)))))))..))))))))).. ( -30.69) >consensus ________UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA ............(..((((((((((........................))))))))))..)....(((((((((..(((((((................)))))))..))))))))).. (-22.43 = -21.83 + -0.60)

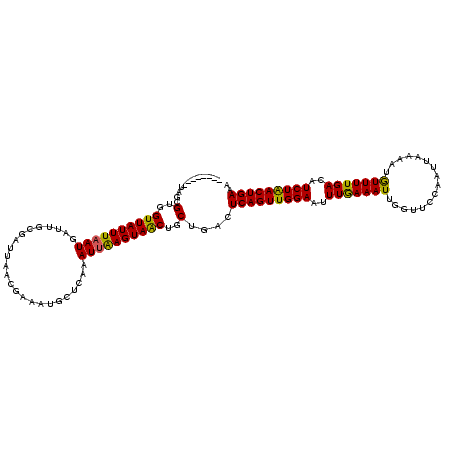
| Location | 20,997,262 – 20,997,374 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -21.05 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.999003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20997262 112 + 23771897 AAUUCUUAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUACGACAGUCCAUAUAUGUCAACUUAAUUA ........((((((((..........(((((((((......(((((....)))))....(((....)))))))))))).......((((.........)))).)))))))). ( -23.20) >DroSec_CAF1 14834 112 + 1 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUCGUUCCAAUUAAAAUAUUUUGACAUCUAACUGAAAUCUGCGACAGCCCAUAUAUGUCAACUUAAUUA ........((((((((....((.((.(((((((((..(..((((...((......))...))))..)..)))))))))..)).))((((.........)))).)))))))). ( -24.30) >DroSim_CAF1 22277 112 + 1 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUGCGACAGCCCAUAUAUGUCAACUUAAUUA ........((((((((....((.((.(((((((((......(((((....)))))....(((....))))))))))))..)).))((((.........)))).)))))))). ( -26.10) >DroEre_CAF1 23583 112 + 1 AAUGCUAAAAUGGAGUAGCUGCUGACUCAGUUGGAAUUUGAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUGACUGAAAUCCACGGCAGUCCAUAUAUGUCAACUUAAUUA .........(((((.(.((((..((.(((((..((..(..((((................))))..)..))..)))))..))..))))).)))))................. ( -23.89) >DroYak_CAF1 21040 112 + 1 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUAAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCCACGACAGUCCAUAUAUGUCAACUUAAUUA ........((((((((..........(((((((((..(((((((................)))))))..))))))))).......((((.........)))).)))))))). ( -21.89) >consensus AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUACGACAGUCCAUAUAUGUCAACUUAAUUA ........((((((((..........(((((((((..(((((((................)))))))..))))))))).......((((.........)))).)))))))). (-21.05 = -20.45 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:40 2006