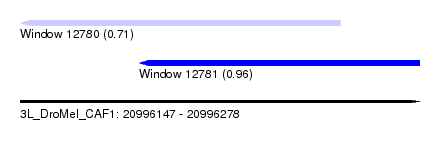
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,996,147 – 20,996,278 |
| Length | 131 |
| Max. P | 0.962688 |

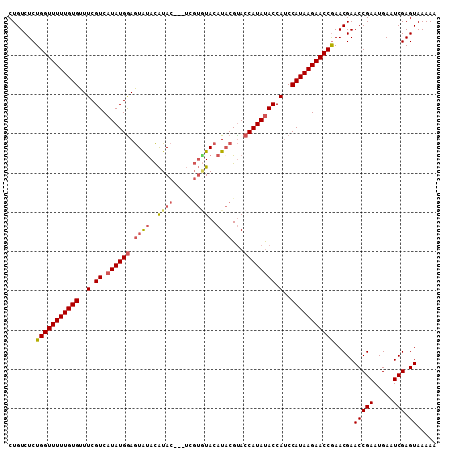
| Location | 20,996,147 – 20,996,252 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -17.94 |
| Energy contribution | -19.25 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20996147 105 - 23771897 CUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUGUACAUAC---UUGUAUACAUACGUACCAUAUACCA-CCAUAAGAACCGAACGAACCGAAUGAAUCGAGUAAAAA .(((.((.((((((((((....((.((((((.(((((((...---.))))))).......))))))))..-.))))))))))))))).(((((.....))).))..... ( -23.80) >DroSec_CAF1 13745 106 - 1 CUGUCUCUGGUUUUUGUGUUUCGUCAUAUGAAGUAUACAUAC---UCGUGUACAUACGUACCAUACACCAUCCAUAAGAACCGAACGAACCGAAUGAAUCGAGUAAAAA .(((.((.((((((((((.(((((...)))))((((((....---..))))))...................))))))))))))))).(((((.....))).))..... ( -20.30) >DroSim_CAF1 21171 106 - 1 CUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUACACAUAC---UCGUGUACAUACGUACCAUAUACCAUCCAUAAGAACCGAACGAACCGAAUGAAUCGAGUAAAAA .(((.((.((((((((((..(.((.((((((.((((((....---..)))))).......)))))))).)..))))))))))))))).(((((.....))).))..... ( -26.00) >DroYak_CAF1 19853 104 - 1 CUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAAUAUAUGUACAUA-----UACAUUCGCACCAUAUACCAUCCAUAAGAACCAAACGAACCGAAUGAAUCGAGUAAAAA .......(((((((((((..(.((.((((((.....(((((....-----))))).....)))))))).)..))))))))))).....(((((.....))).))..... ( -24.30) >consensus CUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUAUACAUAC___UCGUGUACAUACGUACCAUAUACCAUCCAUAAGAACCGAACGAACCGAAUGAAUCGAGUAAAAA .......(((((((((((..(.((.((((((.((((..((((.....))))..))))...)))))))).)..))))))))))).....(((((.....))).))..... (-17.94 = -19.25 + 1.31)


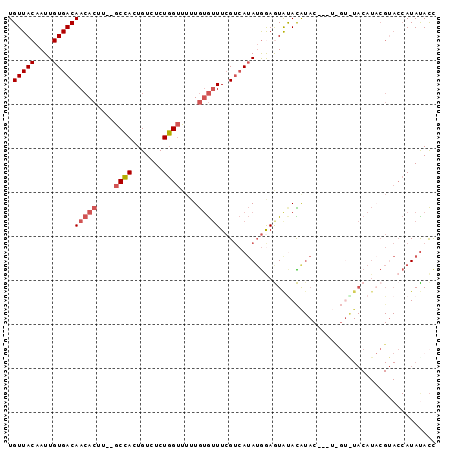
| Location | 20,996,186 – 20,996,278 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -9.00 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20996186 92 - 23771897 UGUUACAAUUGUGACAACACUU--GCCACUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUGUACAUAC---UUGUAUACAUACGUACCAUAUACC ((.(((...(((((((((((..--((((.......))))....)))))..))))))....(((((((...---.)))))))....))).))...... ( -23.00) >DroSec_CAF1 13785 92 - 1 UGUUACAAUUGUGACAACACUU--GCCACUGUCUCUGGUUUUUGUGUUUCGUCAUAUGAAGUAUACAUAC---UCGUGUACAUACGUACCAUACACC ((.(((...(((((((((((..--((((.......))))....)))))..))))))....((((((....---..))))))....))).))...... ( -21.50) >DroSim_CAF1 21211 92 - 1 UGUUACAAUUGUGACAACACUU--GCCACUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUACACAUAC---UCGUGUACAUACGUACCAUAUACC ((.(((...(((((((((((..--((((.......))))....)))))..))))))....((((((....---..))))))....))).))...... ( -23.40) >DroYak_CAF1 19893 90 - 1 UGUUACAAUUGUGACAACACUU--GCCACUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAAUAUAUGUACAUA-----UACAUUCGCACCAUAUACC .(((((....)))))(((((..--((((.......))))....)))))..((.((((((.....(((((....-----))))).....)))))))). ( -20.20) >DroAna_CAF1 11590 74 - 1 UGUUACAAUUGUGACAACACUUUGGCAACUGUCUCUUGACCAUAAAGUUGGAAAAAAAAAAUACAUA-----------------------AUAUAUU ((((((....)))))).........((((((((....))).....))))).................-----------------------....... ( -10.60) >consensus UGUUACAAUUGUGACAACACUU__GCCACUGUCUCUGGUUUUUGUGUUUCGUCAUAUGGAGUAUACAUAC___U_GU_UACAUACGUACCAUAUACC .(((((....)))))(((((....((((.......))))....)))))................................................. ( -9.00 = -9.68 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:37 2006