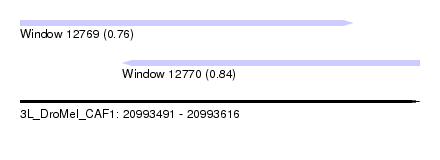
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,993,491 – 20,993,616 |
| Length | 125 |
| Max. P | 0.841909 |

| Location | 20,993,491 – 20,993,595 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -16.82 |
| Energy contribution | -18.02 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20993491 104 + 23771897 ------ACGCUGACCGCCAAAAUCGCUGAUUCAC--UUAUUCAGCAUAAGGUCUGGUGUAAAGGGGGUUUAC-CUUUUGAAUCAUCCUUU-------GUUGGCACUUAAAAUGCAAUUCU ------.........(((((....(((((.....--....))))).(((((..((((.(((((((......)-)))))).))))..))))-------))))))................. ( -27.80) >DroSec_CAF1 11146 103 + 1 ------ACGCUGACCGCCAAAAUCGCUGAAUCAC--UGAUUCAGCAUAAGGCUUGUUGUA-AGGGGGUUUAC-CUUUUGAAUAAUCCUUU-------GUUGGCACUUAAAAUGCAACUCU ------.........(((((....((((((((..--.))))))))..((((.(((((.((-(((((.....)-))))))))))).)))).-------.)))))................. ( -30.20) >DroSim_CAF1 18554 104 + 1 ------ACGCUGACCGCCAAAAUCGCUGAAUCAC--UGAUUCAGCAUAAGGCGUGCUGUAAAGGGGGUUUAC-CUUUUGAAUCAUCCUUU-------GUUGGCACUUAAAAUGCAACUCU ------..((.....(((......((((((((..--.))))))))....)))(((((..((((((((((((.-....)))))).))))))-------...))))).......))...... ( -31.20) >DroEre_CAF1 19656 103 + 1 ------ACGCUGACCGCCAAAAUCGCUGAAUCAC--UGAUUUAGCAUAAUGCCAGUUGUAAAG-GGGCUUAC-CUUUUGAAUCAUCCUUU-------GUUGGCACUUAAAAUGCAGCACU ------..((((............((((((((..--.))))))))....((((((...(((((-((.....)-))))))...((.....)-------))))))).........))))... ( -26.70) >DroYak_CAF1 17328 103 + 1 ------ACGCUGACCGUCAAAAUCGCUGAAUCAC--UGAUUUUGCAUAAAGACAGCUGUAAAG-CGGUUCAC-CUUUUGAAUCAUCCUUU-------GUUGGCACUUAAAAUGCAACACU ------..((..((.(.((((((((.((...)).--)))))))))..((((...(((....))-)((((((.-....))))))...))))-------))..))................. ( -22.50) >DroAna_CAF1 9234 111 + 1 GCAGGAAUUUUGACCGCCAAAAUGGCUGAAUCAUCGUGUUUUCGCUUAAC---------AGAAGGGUUAUGCUUUUUUGAAUCACCUUUUUAGGGGGGGUAGUACUUUAAAUGCAACAUU (((.(((..(((.((.((((((.((.(((......(((....)))....(---------(((((((.....))))))))..))))).)))).)).))..)))...)))...)))...... ( -22.90) >consensus ______ACGCUGACCGCCAAAAUCGCUGAAUCAC__UGAUUCAGCAUAAGGCCUGCUGUAAAGGGGGUUUAC_CUUUUGAAUCAUCCUUU_______GUUGGCACUUAAAAUGCAACACU ...............(((((....((((((((.....))))))))..............((((((((((((......)))))).))))))........)))))................. (-16.82 = -18.02 + 1.20)


| Location | 20,993,523 – 20,993,616 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -16.03 |
| Energy contribution | -16.19 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20993523 93 - 23771897 AAUUCACUUGAAGUGCUUACGAGAAUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAACCCCCUUUACACCAGACCUUAUGCUGA ....((((((((((((...........)))))))))))).((.((((((.((.....(((((.......)))))....))..)))).)).)). ( -20.80) >DroSec_CAF1 11178 92 - 1 AAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUUAUUCAAAAGGUAAACCCCCU-UACAACAAGCCUUAUGCUGA ....((((((((((((..((....)).)))))))))))).((.((((((.........((((.......)))-)........)))).)).)). ( -23.43) >DroSim_CAF1 18586 93 - 1 AAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAACCCCCUUUACAGCACGCCUUAUGCUGA ....((((((((((((..((....)).))))))))))))((......))........(((((.......))))).(((((.......))))). ( -26.30) >DroEre_CAF1 19688 92 - 1 AAAUCACUUAAGCUGCGUACAAGUGCUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAGCCC-CUUUACAACUGGCAUUAUGCUAA .....(((((((.((((((....))).))).)))))))((((......((....)).(((((......)-)))).....)))).......... ( -19.50) >DroYak_CAF1 17360 92 - 1 AAAUCACUUGAAGUGCUUACAAGUGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUGAACCG-CUUUACAGCUGUCUUUAUGCAAA ....((((((((((((..((....)).))))))))))))......((((((.........((....))(-((....))).))))))....... ( -23.10) >consensus AAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAACCCCCUUUACAACAGGCCUUAUGCUGA ....((((((((((((...........)))))))))))).......((.((((........(((((.....)))))........)))).)).. (-16.03 = -16.19 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:27 2006