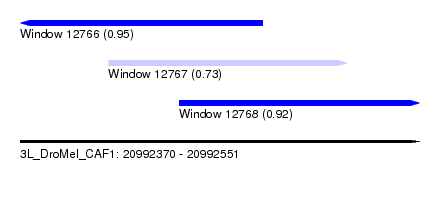
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,992,370 – 20,992,551 |
| Length | 181 |
| Max. P | 0.949157 |

| Location | 20,992,370 – 20,992,480 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.20 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
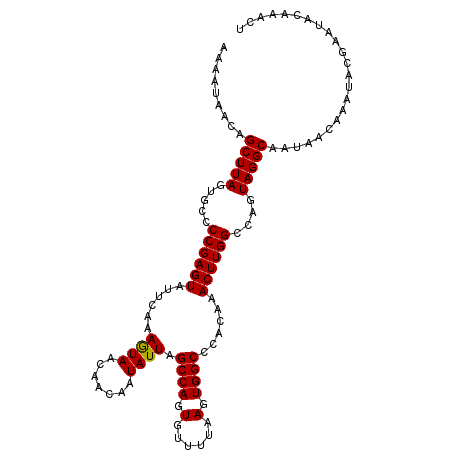
>3L_DroMel_CAF1 20992370 110 - 23771897 UGUUACUUUGAAUACUCGGGGCACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUGAAAUAACAUGCAUACAGAAAACUCUAUCCCUUAACCACU .................((((......(((((((((((....(((((((........)))))))......))))))))).))....(((....))).))))......... ( -21.40) >DroSec_CAF1 10034 97 - 1 UUUUACUUUGAAUACUCGGGGCACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUAAAAUAACAUGCAUA-------------CCCUUAACCACU .................((((...((.(((((((((((....(((((((........)))))))......))))))))).)).))-------------))))........ ( -19.00) >DroSim_CAF1 17445 90 - 1 UGUUACUUUGAAUACUCGGGGCACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUAAAAUAAU--------------------CCCUUAACCACU .....((((((....))))))...((((..((((((((....(((((((........)))))))......))))))))--------------------..))))...... ( -15.10) >DroEre_CAF1 18517 110 - 1 UGUUAUUUCGAAUACUCGGGGUACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUAAAAUAUCAUGCAUACAGAAAACUUGAUCCCUUGACCACU .((((....((....))(((((.....((((.((((((....(((((((........)))))))......)))))).)).))...(((.....)))))))).)))).... ( -18.20) >DroYak_CAF1 16196 110 - 1 UGUUAUUUUGAAUACUCGGGGUACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUAAAAUAACAUGCAUACAGAAAACUUGUUCCCUUGACCACU ...............((((((......(((((((((((....(((((((........)))))))......))))))))).))..((((.....))))..))))))..... ( -21.50) >consensus UGUUACUUUGAAUACUCGGGGCACUAAGCUGUUAUUUUCCACUUGAUCUGAUUUCAAAGAUCAAAAUUUUAAAAUAACAUGCAUACAGAAAACU___UCCCUUAACCACU .................(((((.....))(((((((((....(((((((........)))))))......)))))))))...................)))......... (-15.90 = -15.74 + -0.16)

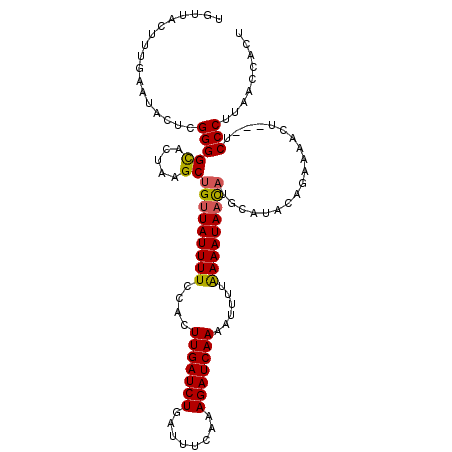
| Location | 20,992,410 – 20,992,518 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 98.15 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20992410 108 + 23771897 AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((((((((.....)))))..)))).............((((.(......).))))))))....... ( -21.60) >DroSec_CAF1 10061 108 + 1 AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAAAAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((((((((.....)))))..)))).............((((.(......).))))))))....... ( -21.60) >DroSim_CAF1 17465 108 + 1 AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((((((((.....)))))..)))).............((((.(......).))))))))....... ( -21.60) >DroEre_CAF1 18557 108 + 1 AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUACCCCGAGUAUUCGAAAUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((..(....)..)))......................((((.(......).))))))))....... ( -19.90) >DroYak_CAF1 16236 108 + 1 AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUACCCCGAGUAUUCAAAAUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((..(....)..)))......................((((.(......).))))))))....... ( -19.90) >consensus AAAAUUUUGAUCUUUGAAAUCAGAUCAAGUGGAAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUG ......(((((((........)))))))((((.........(((..(....)..)))......................((((.(......).))))))))....... (-19.90 = -19.90 + 0.00)


| Location | 20,992,442 – 20,992,551 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -16.98 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20992442 109 + 23771897 AAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAGUAGGCAAUAACAAAUACGAAUACAAACU .........(((((.((..((((((.......((....)).......((((.(......).))))......)))))).)).)))))....................... ( -20.70) >DroSec_CAF1 10093 109 + 1 AAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAAAAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAGUAGGCAAUAACAAAUACGAAUACAAACU .........(((((.((..((((((.......((....)).......((((.(......).))))......)))))).)).)))))....................... ( -20.70) >DroSim_CAF1 17497 109 + 1 AAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAGUAGGCAAUAACAAAUACGAAUACAAACU .........(((((.((..((((((.......((....)).......((((.(......).))))......)))))).)).)))))....................... ( -20.70) >DroEre_CAF1 18589 109 + 1 AAAAUAACAGCUUAGUACCCCGAGUAUUCGAAAUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAAUAGGCAAUAACAAAUACAAGUGCAAACU .........(((((.....((((((......((((.......)))).((((.(......).))))......))))))....)))))....................... ( -17.40) >DroYak_CAF1 16268 109 + 1 AAAAUAACAGCUUAGUACCCCGAGUAUUCAAAAUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAAUAGGCAAUAACAAAUACAAAUGCAAACU .........(((((.....((((((......((((.......)))).((((.(......).))))......))))))....)))))....................... ( -17.40) >consensus AAAAUAACAGCUUAGUGCCCCGAGUAUUCAAAGUAACAACAAUAUUAGCCAGUGUUUUAAGUGGCCCACAAACUUGGCCAGUAGGCAAUAACAAAUACGAAUACAAACU .........(((((.....((((((......((((.......)))).((((.(......).))))......))))))....)))))....................... (-16.98 = -16.74 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:25 2006