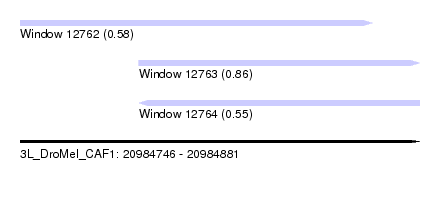
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,984,746 – 20,984,881 |
| Length | 135 |
| Max. P | 0.857612 |

| Location | 20,984,746 – 20,984,865 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -20.49 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.99 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
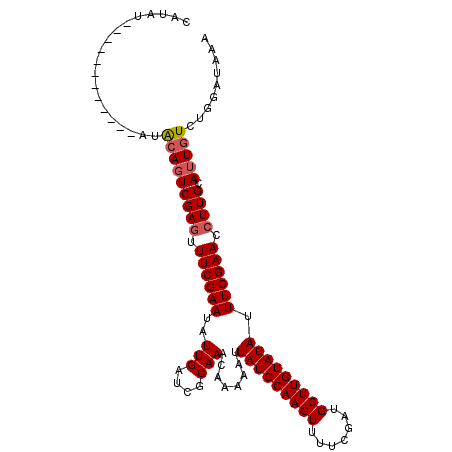
>3L_DroMel_CAF1 20984746 119 + 23771897 CGCUUUACGAAACAAAUUUAAUAAACAAUAAUUGUUAAUAUUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAU-UUUUGUUUGCGAUCAAUAUUCGAAU .......((((......(((((((.......)))))))((((.((((((((((((((((.(((((............)))))..)))))))...-...)))))).))).))))))))... ( -23.20) >DroSec_CAF1 2425 119 + 1 CGCUUUACAAAACAUAUUUAAUAAACAAUAAUUGUUAAUGUUUAUUCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAU-UUUUGUUUGCGAUCAAUAUUCGAAA (((...((((((.......((((((((.((.....)).))))))))......(((((((.(((((............)))))..)))))))...-))))))..))).............. ( -22.80) >DroSim_CAF1 9823 119 + 1 CGCUUUACAAAACAUAUUUAAUAAACAAUAAUUGUUAAUGUUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAU-UUUUGUUUGCGAUCAAUAUUCGAAA (((...((((((........(((((((.((.....)).))))))).......(((((((.(((((............)))))..)))))))...-))))))..))).............. ( -22.00) >DroEre_CAF1 10845 95 + 1 CUCUAUACAAAACAUAUUUAAUAAACGAUAAUUGUUAAUAUUUAUCCAGACAAUGGAAGG-------------------------UUGCAUAACUUUUUAUUUGCGAUCAAUAUUCGGAU .........................(((....((((........((((.....)))).((-------------------------(((((............))))))))))).)))... ( -13.80) >DroYak_CAF1 8535 120 + 1 CGCUUUACAAAACAUAUUUAAUAAACGAUAAUUGUUAAUAUAUAUCCAGACAAUGCAAGGUUCAAAAUAAACCAACAUCGGAAGCUUGCAUAAUUUUUUGUUUGCGAUCAAUAUUCGAAU (((...((((((.(((((((((((.......))))))).)))).........(((((((.(((.................))).)))))))....))))))..))).............. ( -20.63) >consensus CGCUUUACAAAACAUAUUUAAUAAACAAUAAUUGUUAAUAUUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAU_UUUUGUUUGCGAUCAAUAUUCGAAU .......................(((((...)))))((((((.((((((((((((((((.(((((............)))))..))))))).......)))))).))).))))))..... (-13.50 = -13.99 + 0.49)


| Location | 20,984,786 – 20,984,881 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20984786 95 + 23771897 UUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAUUUUUGUUUGCGAUCAAUAUUCGAAUAUCGACUCCAA------------AACUU ...((((((((((((((((.(((((............)))))..)))))))......)))))).)))......(((.....)))......------------..... ( -18.10) >DroSec_CAF1 2465 95 + 1 UUUAUUCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAUUUUUGUUUGCGAUCAAUAUUCGAAACUCGACUGUAU------------AUAUG ..(((.(((...(((((((.(((((............)))))..))))))).......((((.(((.......)))))))....))).))------------).... ( -18.00) >DroSim_CAF1 9863 107 + 1 UUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAUUUUUGUUUGCGAUCAAUAUUCGAAACUCGAAUGUAUUUAGCUAUACUUAUAAG ...((((((((((((((((.(((((............)))))..)))))))......)))))).)))..(((((((.....)))))))................... ( -22.80) >consensus UUUAUCCAGACAAUGCAAGGUUCGAAAUAUACAAGCAUCGAAAACUUGCAUAAUUUUUGUUUGCGAUCAAUAUUCGAAACUCGACUGUAU____________AUAUG ...((((((((((((((((.(((((............)))))..)))))))......)))))).)))......(((.....)))....................... (-17.45 = -17.23 + -0.22)



| Location | 20,984,786 – 20,984,881 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -17.32 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20984786 95 - 23771897 AAGUU------------UUGGAGUCGAUAUUCGAAUAUUGAUCGCAAACAAAAAUUAUGCAAGUUUUCGAUGCUUGUAUAUUUCGAACCUUGCAUUGUCUGGAUAAA ....(------------(..((((((((((....)))))))).((((........(((((((((.......))))))))).........))))....))..)).... ( -20.53) >DroSec_CAF1 2465 95 - 1 CAUAU------------AUACAGUCGAGUUUCGAAUAUUGAUCGCAAACAAAAAUUAUGCAAGUUUUCGAUGCUUGUAUAUUUCGAACCUUGCAUUGUCUGAAUAAA ....(------------(.(((((((((.((((((..(((....)))........(((((((((.......))))))))).)))))).)))).))))).))...... ( -21.20) >DroSim_CAF1 9863 107 - 1 CUUAUAAGUAUAGCUAAAUACAUUCGAGUUUCGAAUAUUGAUCGCAAACAAAAAUUAUGCAAGUUUUCGAUGCUUGUAUAUUUCGAACCUUGCAUUGUCUGGAUAAA .......((((......))))(((((.....)))))....(((.((.(((......(((((((..(((((............))))).)))))))))).)))))... ( -19.50) >consensus CAUAU____________AUACAGUCGAGUUUCGAAUAUUGAUCGCAAACAAAAAUUAUGCAAGUUUUCGAUGCUUGUAUAUUUCGAACCUUGCAUUGUCUGGAUAAA ...................(((((((((.((((((..(((....)))........(((((((((.......))))))))).)))))).)))).)))))......... (-17.32 = -18.10 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:22 2006